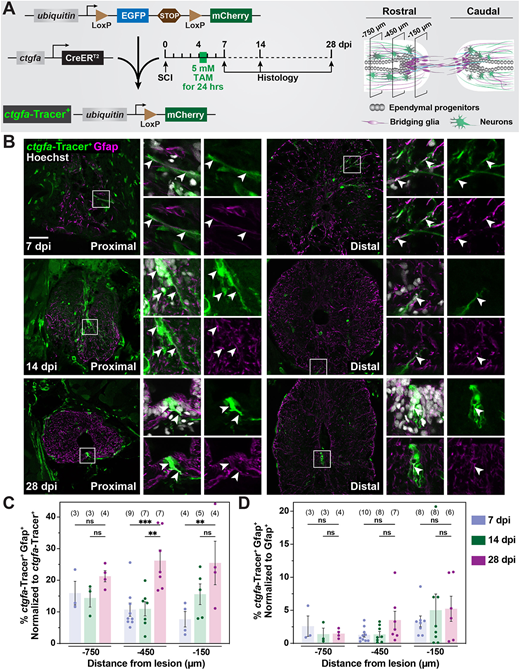
Fig. 1 Contribution of ctgfa+ cells to regenerating glia during SC repair. (A) Experimental timeline to evaluate the contribution of ctgfa+ cells after SCI. ctgfa-Tracer animals refer to the compound transgenic line −5.5Kb-ctgfa:CreERT2; ubiquitin:loxP-GFP-STOP-loxP-mCherry. ctgfa-Tracer fish were subjected to complete SC transection and tamoxifen (TAM)-mediated recombination at 4 dpi to enable permanent mCherry labeling in ctgfa+ and ctgfa+-derived cells. SC tissues were collected at 7, 14 and 28 dpi. Schematic of SC tissue illustrates cross-sections at 150, 450 and 750 µm rostral to the lesion that were analyzed for histological examination. The 7 dpi time point was used to assess recombination following TAM treatment. The 14 and 28 dpi time points were used to trace the fates of ctgfa+-derived cells. (B) Immunostaining for mCherry (green), Gfap (magenta), and nuclear Hoechst (gray) at 7, 14 and 28 dpi. mCherry expression, referred to as ctgfa-Tracer+, is used to trace the fates of ctgfa-expressing cells following TAM-inducible recombination. SC sections from TAM-treated ctgfa-Tracer (Cre+) animals are shown. Cross-sections 150 µm (proximal) or 450 µm (distal) rostral from the lesion site are shown. High-magnification insets show select ctgfa-Tracer+ cells in triple-, double- or single-channel views. Arrowheads indicate ctgfa-Tracer+ cells. (C,D) Quantification of ctgfa-Tracer+ and Gfap+ colocalization at 7, 14 and 28 dpi. SC cross-sections 150, 450, and 750 µm rostral to the lesion were analyzed. ctgfa-Tracer+ and Gfap+ fluorescence were quantified. For each section, ctgfa-Tracer+ Gfap+ fluorescence was normalized to total ctgfa-Tracer+ in C and to total Gfap+ in D. Dots indicate individual animals and sample sizes are indicated in parentheses. Error bars represent s.e.m. ***P<0.001; **P<0.01 (two-way ANOVA). ns, not significant (P>0.05). Scale bar: 50 µm.
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development