Fig. 4
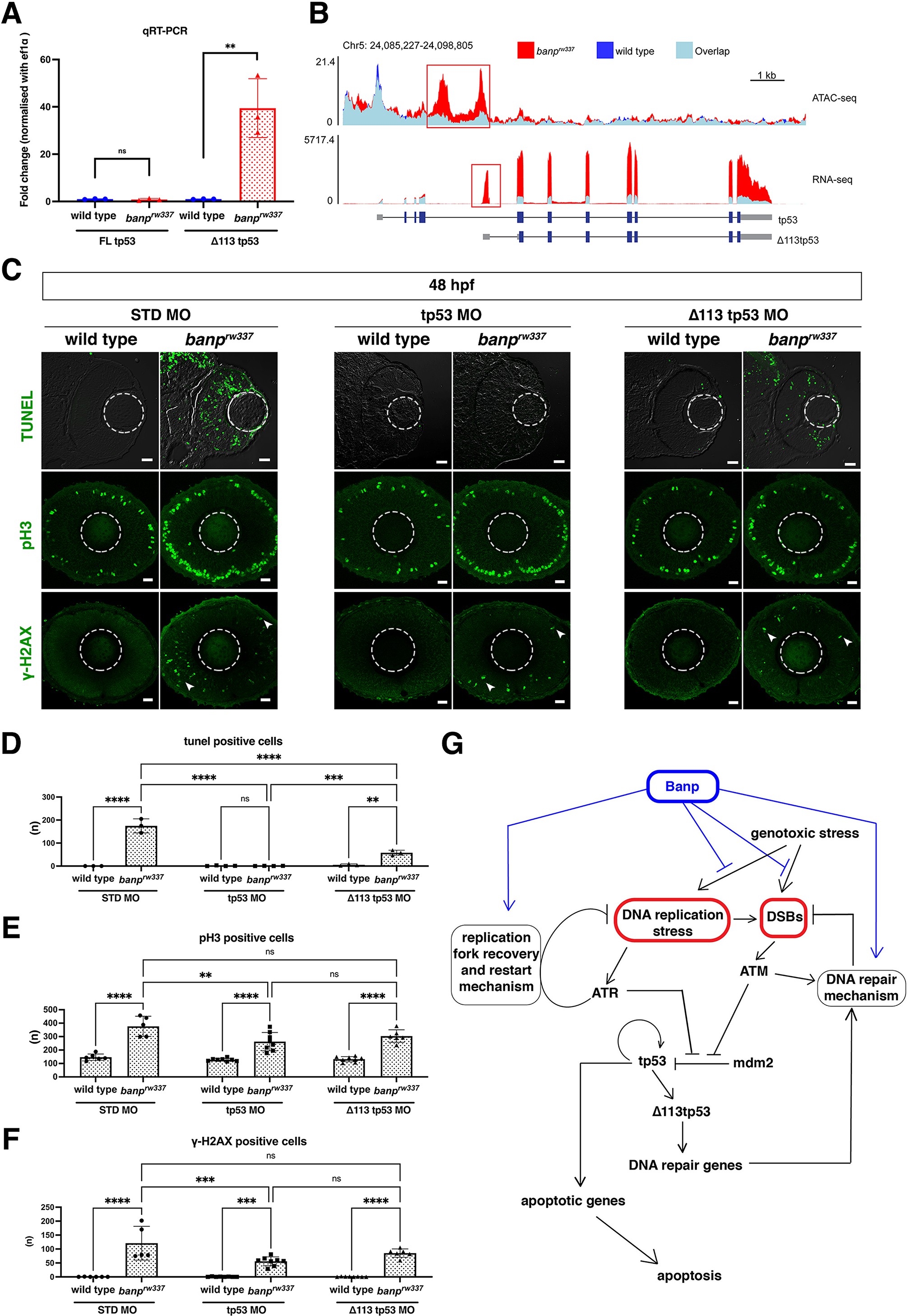
(A) Quantitative RT-PCR-based validation of FL tp53 and ∆113tp53 mRNA expression at 48 hpf. ∆113tp53 mRNA expression is markedly higher in banprw337 mutants than in wild-type siblings, whereas FL tp53 mRNA is not significantly different between mutants and wild-type siblings. Unpaired t-test (two-tailed), Mean ± SD. [n=3, p**<0.01, ns (not significant)]. (B) ATAC-seq- and RNA-seq-based validation of chromatin accessibility (upper panel) and transcription level (bottom panel) of the genomic region covering both tp53 and ∆113tp53 transcription units. Data of wild-type and banprw337 mutants are indicated in blue and red, respectively. Light blue indicates overlapping level. The red openbox in ATAC-seq shows open chromatin in banprw337 mutants in intron 4, signifying differential transcription of ∆113tp53 using an internal promoter. (C) TUNEL (top), anti-pH3 antibody labeling (middle), and anti-γ-H2AX antibody labeling (bottom) of wild-type and banprw337 mutant retinas injected with STD MO, tp53 MO, and ∆113tp53 MO. TUNEL data of 48 hpf wild-type sibling and banprw337 mutant retinas injected with STD MO and tp53 MO are shared with those shown in Figure 3G. tp53 MO-mediated knockdown completely rescues apoptosis in banprw337 mutant retinas at 48 hpf. However, ∆113tp53 MO-mediated knockdown mildly inhibits apoptosis in banprw337 mutant retinas. Neither tp53 MO nor ∆113tp53 MO effectively rescued mitotic cell accumulation and accumulation of γ-H2AX-positive cells (white arrowheads) in banprw337 mutant retinas. Scale bars: 20 μm. (D–F) Histogram of the number of TUNEL-positive cells (D), pH3-positive cells (E), and γ-H2AX-positive cells (F) per retinal section in wild-type and banprw337 mutants injected with STD-MO, tp53 MO, and ∆113tp53 MO. TUNEL data of 48 hpf wild-type sibling and banprw337 mutant retinas injected with STD MO and tp53 MO are shared with those shown in Figure 3H. Two-way ANOVA with Tukey’s multiple comparisons test, Mean ± SD. [n=in graph, p*<0.05, p**<0.01, p***<0.001, p****<0.0001, ns (not significant)]. (G) Possible role of Banp in tp53-mediated DNA damage response pathway. DNA replication stress activates ATR, which subsequently promotes DNA replication fork recovery. Failure of stalled fork recovery causes DSBs, which activate ATM-dependent DNA damage repair. ATR and ATM inhibit Mdm2-mediated tp53 degradation. Stabilized tp53 initially promotes transcription of the ∆113tp53 isoform, which activates cell-cycle arrest genes and promotes DNA damage repair. Chronic activation of tp53 promotes transcription of apoptotic genes to induce apoptosis. Banp may normally suppress DNA replication stress by promoting DNA replication fork recovery or DNA damage repair. Banp may suppress genotoxic stress-mediated activation of DNA replication stress and DSB formation.
Banp is required for integrity of DNA replication and DNA damage repair.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Elife