FIGURE 1
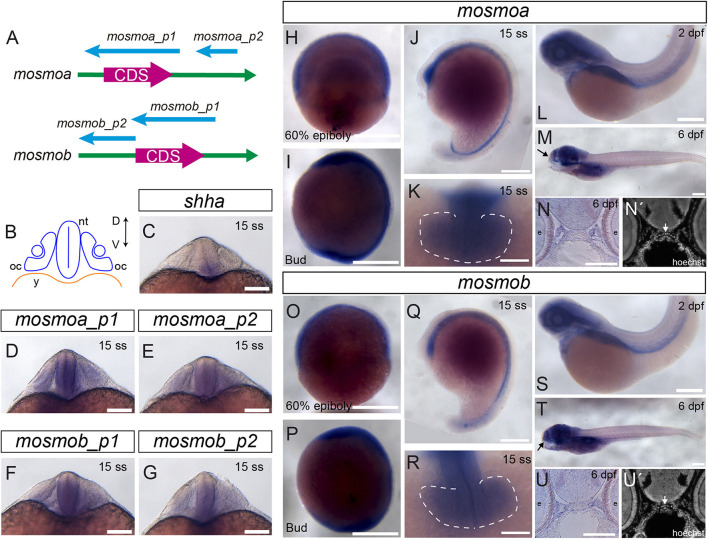
Figure 1. Mosmo paralogs show a largely overlapping distribution in zebrafish. (A) Schematic representation of mosmoa and mosmob mRNAs and probes used for in situ hybridization (ISH). p1, probe #1. p2, probe #2. (B) Schematic representation of a frontal section of a zebrafish embryo at 20 hpf at the level of the optic cup. Expression pattern of shha (C), mosmoa (D,E) and mosmob (F,G) at 20 hpf. Note that the two probes for mosmoa and mosmob show an identical distribution. Note also that both paralogs have an overlapping distribution in the ventral neural tube but more dorsally extended than that of shha. (H,U) Expression pattern of embryos hybridized with mosmoa and mosmob at 60% epiboly (H,O), bud (I,P) and 15 ss stage (J,K,Q,R), 2 dpf (L,S) as well as at 6 dpf (M,T). Note that at 60% epiboly and bud stage the expression of both genes is localized along the ventral anterior-posterior axis of the embryos. At 15 ss, a low level expression of both mosmoa and mosmob is detected in the optic vesicles (K,R, dashed line) and along the ventral neural tube from the diencephalon to the tail bud (J,Q). At 2 and 6 dpf, the expression of both genes localizes to the head region. The eyes were removed in panels (M,T). Frontal sections of 6 dpf embryos hybridized in toto for mosmoa (N) or mosmob (U) and counterstained with Hoechst (N′,U′). ISH signal for both paralogs localizes around the ethmoid cartilage (N,N′,U,U′ white arrows). D, dorsal; V, ventral; oc, optic cup; nt, neural tube; and y, yolk. Scale bars: 200 μM.