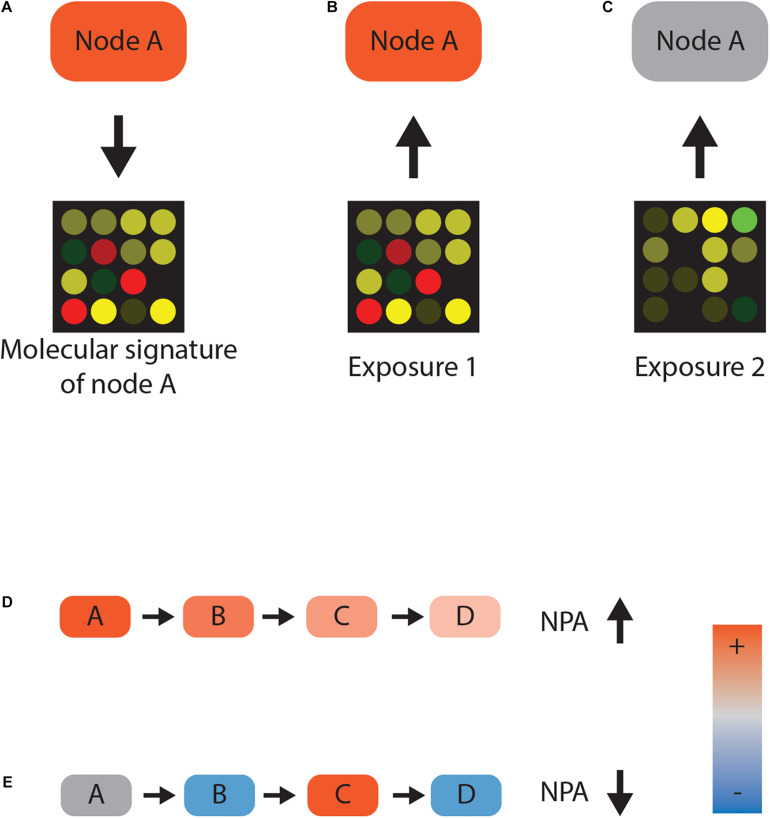
FIGURE 1 Network scoring. (A–C) Reverse causal reasoning paradigm. Gene expression data following activation of protein A can be used as a molecular signature for the activity of that protein A. Any given transcriptomic dataset—for example, from chemically exposed samples—can now be used to infer the activity of protein A. Closely matching gene expression values indicate that protein A was active (B), whereas disparate gene expression is not consistent with the activity (C). (D,E) Network perturbation amplitude (NPA) calculation. If nodes A, B, and C positively regulate node D and their activities are inferred to be high, there is a high consistency between gene expression and network topology. High consistency leads to a high NPA value (D). Conversely, if node activity and the topology of the network contradict each other, the NPA value decreases (E).
Image
Figure Caption
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Front Genet