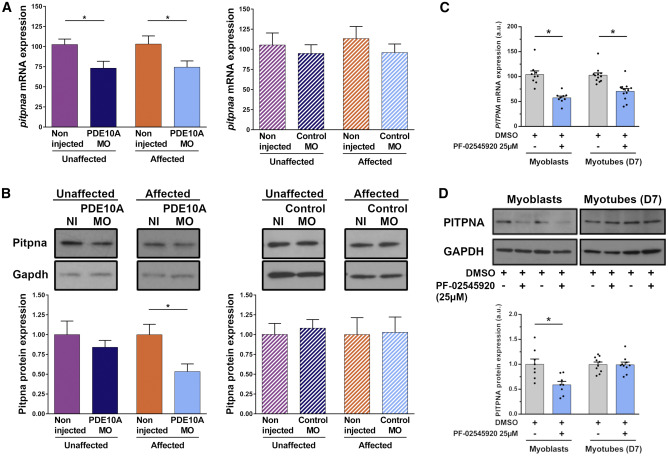
Fig. 7 PDE10A Inhibition Leads to Reduction of PITPNA Expression in Dystrophin-Deficient Zebrafish and DMD Patient-Derived Myoblasts Pairs of sapje-like+/− zebrafish were mated and pde10a morpholino or standard control morpholino (negative control) was injected in one-cell-stage progeny embryos. At 4 dpf, a birefringence assay was performed and unaffected (normal muscle birefringence phenotype) as well as affected larvae (abnormal muscle birefringence phenotype) were collected. In each experiment, morphant zebrafish were compared to non-injected siblings. (A) Total mRNA was extracted from 10 (n = 10) zebrafish per biological replicate and TaqMan qRT-PCR was performed. Bar graphs show pitpna mRNA expression normalized to hprt1. In each experiment, the replicate was run three times and at least five (N = 5) independent experiments were performed. Statistical differences between groups are presented as follows: ∗p < 0.05 (t test, ±SEM). (B) Total proteins were extracted from 20 (n = 20) zebrafish per biological replicate and western blotting was performed. Upper panel: representative western blot images from Pitpna and Gapdh antibodies. Lower panel: bar graph showing Pitpna protein expression normalized to Gapdh. At least four (N = 4) independent experiments were performed. Statistical differences between groups are presented as follows: ∗p < 0.05 (t test, ±SEM). (C) In a primary culture of CD56-positive myoblasts and myotubes (at day 6 of differentiation) from DMD patients, 25 μM PDE10A inhibitor PF-02545920 or 0.25% DMSO (control) was applied for 24 h. Total mRNA was extracted and TaqMan qRT-PCR was performed. Bar graph shows PITPNA mRNA expression normalized to GAPDH. (D) Total proteins were extracted and western blotting was performed. Upper panel: representative western blot images with PITPNA and GAPDH antibodies. Lower panel: bar graph showing PITPNA protein expression normalized to GAPDH. In each experiment, two DMD patient cell lines were used, and replicates were run three times in at least three (N = 3) independent experiments. Statistical differences between groups are presented as follows: ∗p < 0.05 (t test, ±SEM).
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Mol. Ther.