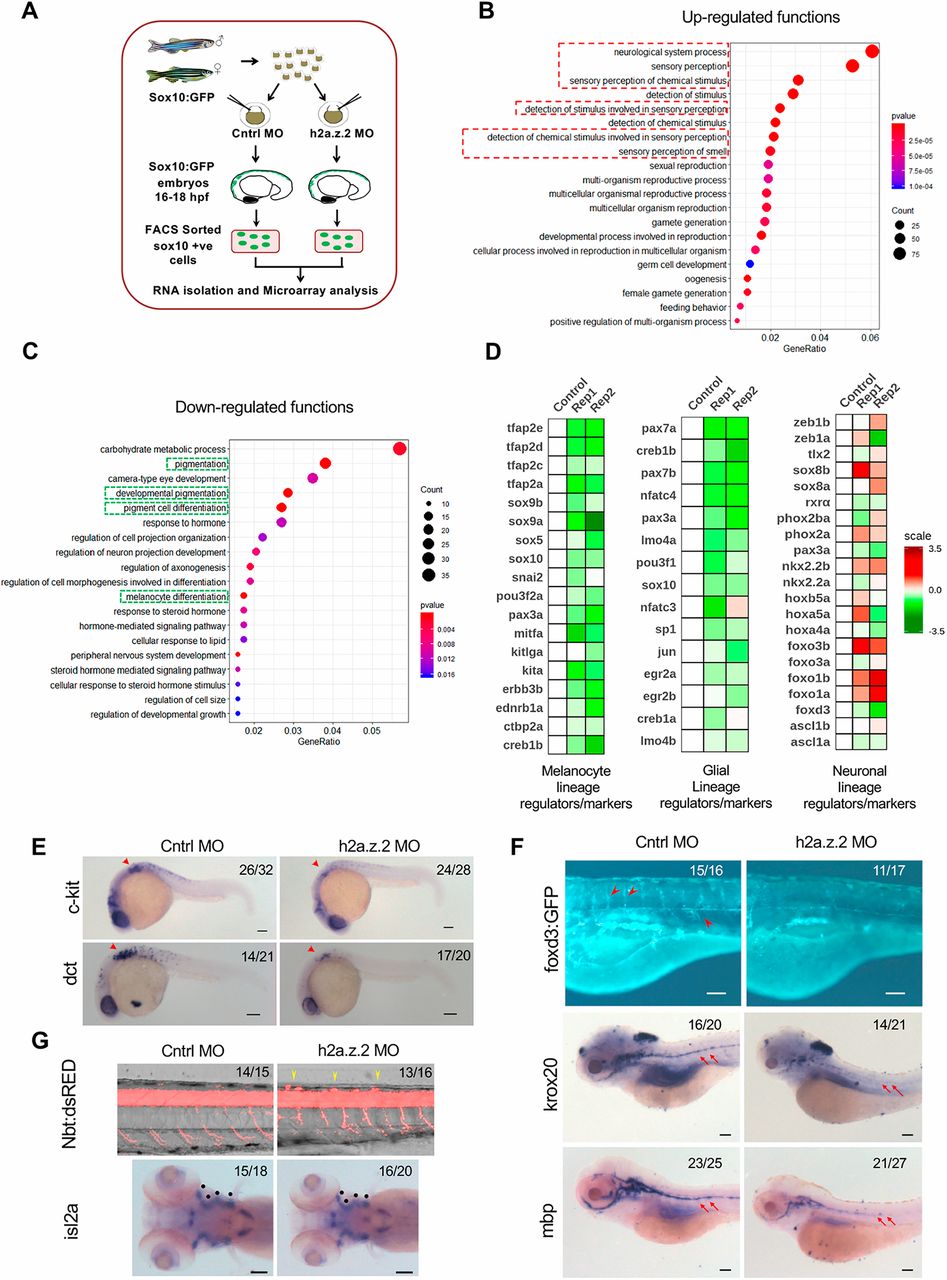
Fig. 2
Fig. 2 H2a.z.2 alters the neural crest gene regulatory network and decreases the melanocyte and glial footprint. (A) Schematic design of the microarray experiment. (B,C) Bubble plots representing gene ontology functions of differentially regulated genes with a log2 fold change ≤−0.6 (downregulated, B) and ≥0.6 (upregulated, C) upon the silencing of Z2 in sox10+ cells. (D) Heatmaps representing differential expression of key transcription factors and lineage markers in sox10+ cells of Z2 MO compared with the control across melanocyte, glial and neuronal lineages. (E) WISH-based expression pattern of melanocyte markers c-kit and dct at 24 hpf. Arrowhead represents vagal melanocytes. (F) Top: Tg(foxd3:GFP) labelling of glial cells that mark undifferentiated glia. Red arrowheads show cell population of interest. Middle and bottom: WISH of glial markers krox20 and mbp at 5 dpf. Red arrows indicate the glial lateral line. (G) Top: Tg(nbt:dsRed) labelling of neurons in the spinal cord bundle (lateral view). Yellow arrowheads mark the ectopic neurons. Bottom: WISH of the cranial motor neuron isl2a. Black dots represent motor neurons that arise from neural crest cells. The numbers in the top right corners of E, F and G indicate number of embryos with observed phenotypes/total number of embryos analysed. Scale bars: 100 μm.