Fig. 8
Fig. 8
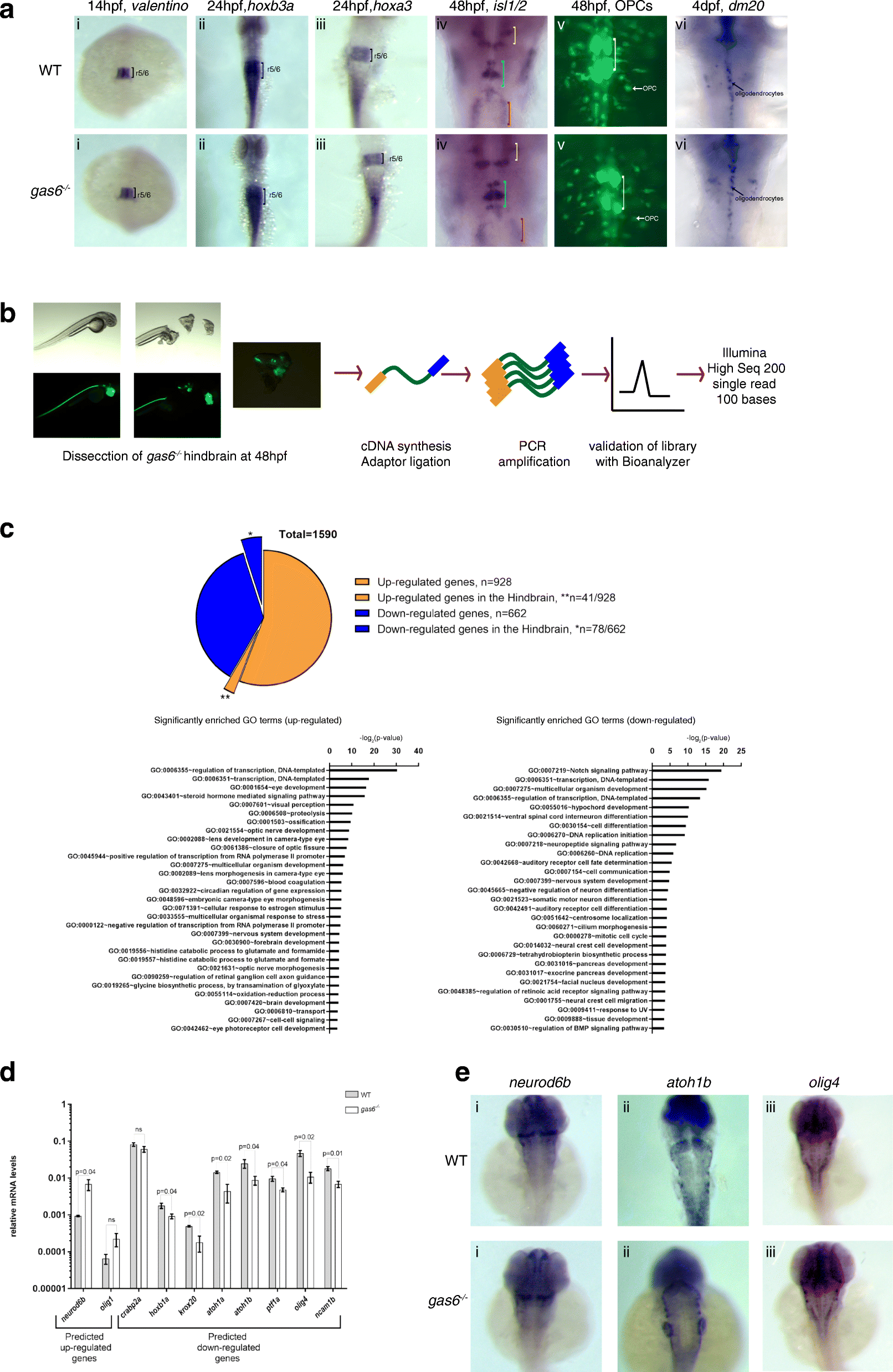
gas6 may only have subtle roles in caudal hindbrain development. a WT and gas6 mutant embryos were assayed for expression of valentino (r5/r6) (i), hoxb3a (r5-spinal cord) (ii), hoxa3 (r5/r6) (iii), islet1 (cranial nerves)(iv) and dm20 (oligodendrocyte marker) (vi) by ISH, as well as for the presence of OPCs and abducens neurons by crossing to the Tg (olig2:EGFP) vu12 line (v). In column (iv), yellow brackets mark cranial nerve V, blue brackets mark cranial nerve VII and red brackets mark cranial nerve X. White brackets indicate the presence of abducens (cranial nerve VI) in column (v). b Schemes showing RNA-seq library synthesis. Hindbrain tissue was dissected from 48 hpf gas6 mutant embryos in the olig2:eGFP background. Total RNA was collected from pools of hindbrain tissue and was used in library synthesis following the TruSeq Stranded mRNA Library Prep Kit (Illumina) protocol. c 1590 differentially expressed genes were identified from RNA-Seq where 41 out of the 928 up-regulated genes and 78 out of the 662 down-regulated genes were expressed in the hindbrain. GO terms related to Biological Processes were identified in both up-regulated and down-regulated genes using DAVID. d A subset of differentially expressed genes was validated via qPCR from independently collected hindbrain tissue samples. e ISH analysis of representative differentially expressed hindbrain genes (i) neurod6b, (ii) atoh1b and (iii) olig4 show no detectable change in expression pattern in gas6 mutant embryos