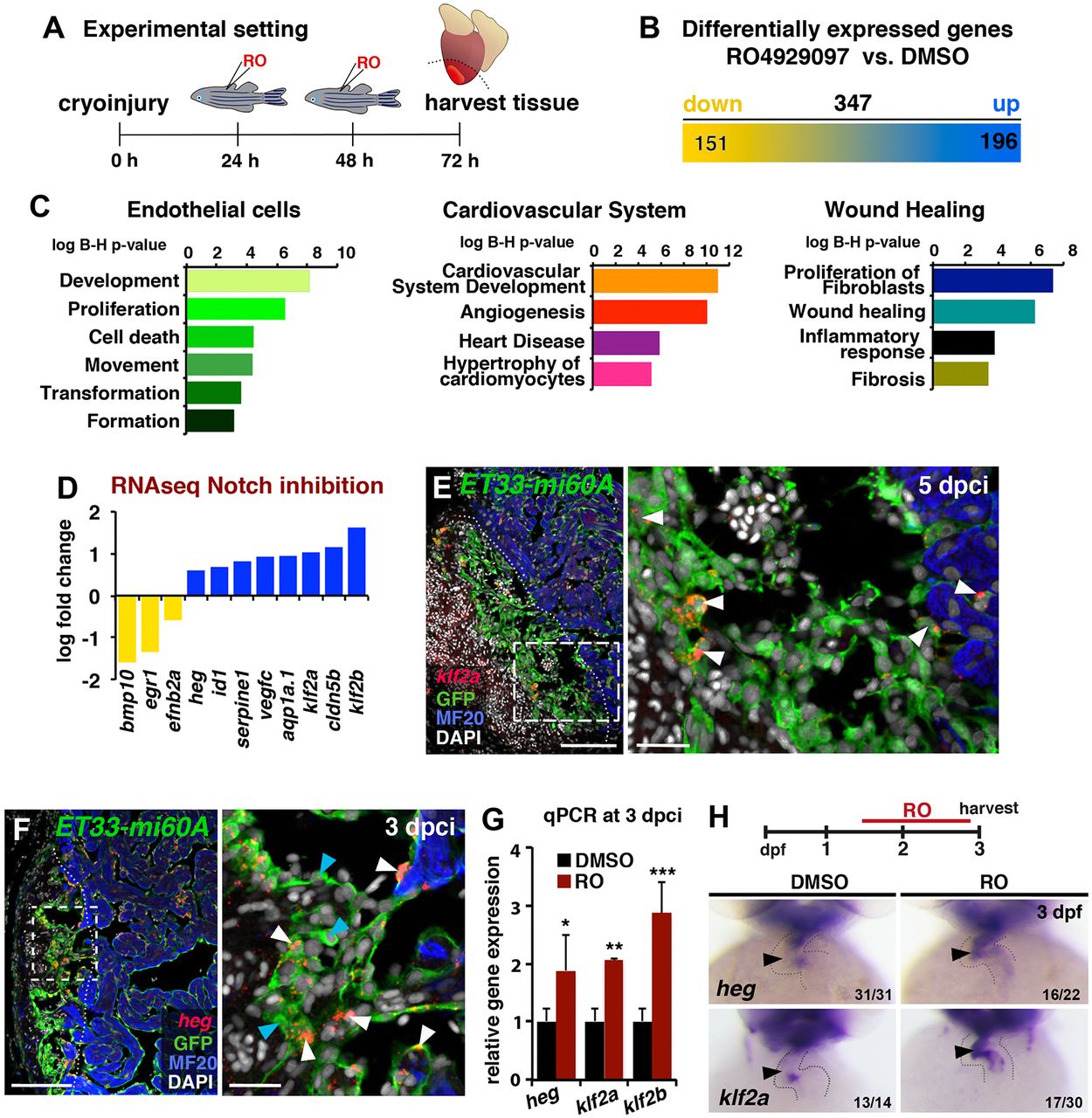
Fig. 4
Fig. 4
Notch inhibition affects developmental and injury-related endocardial/endothelial genes. (A) RO injection and RNA extraction procedure from the ventricular apex (below the dotted line). (B) RNA-seq detected 347 genes differentially expressed in injured hearts after RO treatment (fold change >0.5, P<0.05). (C) Ingenuity-based RNA-seq data analysis. The charts represent selected Ingenuity categories affected. (D) RNA-seq revealed differential cardiac expression of endothelial genes at 3 dpci in RO- or DMSO-treated fish. Yellow, downregulated; blue, upregulated. (E,F) FISH against klf2a (E) or heg (F) combined with IHC. Mosaic expression of both genes was observed in wound and wound-adjacent GFP+ endocardial cells (arrowheads). Boxed areas are magnified on the right. (G) qPCR analysis of heg, klf2a and klf2b expression in injured hearts. Mean±s.d.; t-test, *P<0.05, **P<0.01, **P<0.005. (H) ISH in embryos at 3 days post fertilisation (dpf). RO treatment (regime indicated at top) expanded ventricular expression of heg and klf2a (arrowhead). Dotted lines delineate injury site. The number of embryos displaying the illustrated phenotype among the total number examined is indicated. Scale bars: 100 µm in E,F; 20 µm in magnified views in E,F.