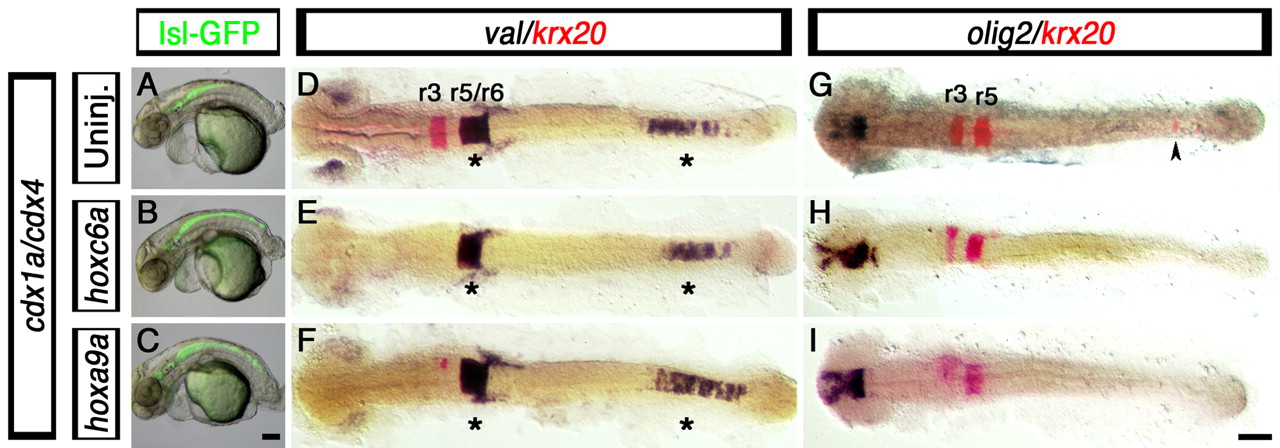
Fig. 7 Hindbrain and spinal cord defects associated with loss of Cdx function are not rescued by posterior Hox gene overexpression. (A-I) Branchial motor neuron distribution (A-C, GFP-positive cells), hindbrain markers krx20 (D-I, red) and val (D-F, purple), and spinal cord oligodendtrocyte marker olig2 (G-I, purple staining), in cdx1a/cdx4-deficient zebrafish embryos injected with 25 pg of hoxc6a and hoxa9a mRNA. (A-C) At 50 hpf, control and hoxc6a and hoxa9a mRNA-injected cdx1a/cdx4-deficient, isl1:GFP transgenic embryos show GFP-positive branchiomotor neurons throughout the posterior CNS. (D-I) At the 20-somite stage (19 hpf), hoxc6a and hoxa9a overexpression in cdx1a/cdx4-deficient embryos results in reduced krx20 expression in r3 and r5 and its loss in the posterior CNS (E,F,H,I, red; caudal expression indicated with an arrowhead), as compared with uninjected controls (D,G and Fig. 2). (D-F) val expression is maintained in r5/6 and posterior CNS of cdx1a/cdx4-deficient embryos overexpressing posterior Hox genes (see Fig. 2 for wild-type control). (G,I) Posterior Hox gene overexpression does not rescue spinal cord olig2 expression in cdx1a/cdx4-deficient embryos (see Fig. 1 for wild-type control). Embryos shown in lateral (A-C) or dorsal (D-I) views, anterior to the left. Asterisk indicates val expression in hindbrain and posterior CNS. Arrowhead indicates ectopic krx20 expression in the posterior CNS. A minimum of 15 embryos in three independent experiments were analyzed, with more than 90% of embryos displaying the phenotypes shown. Scale bars: 100 µm.
Image
Figure Caption
Figure Data
Acknowledgments
This image is the copyrighted work of the attributed author or publisher, and
ZFIN has permission only to display this image to its users.
Additional permissions should be obtained from the applicable author or publisher of the image.
Full text @ Development