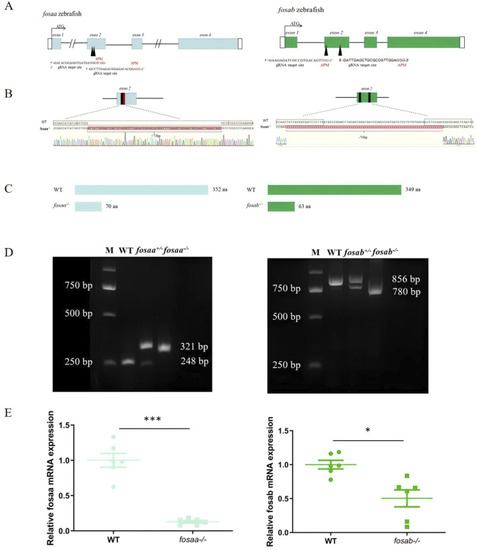
Generation of fosaa−/− and fosab−/− zebrafish using CRISPR/Cas9. (A): Schematic representation of gene structure for fosaa and fosab. The open reading frame (ORF) of fosaa and fosab are respectively indicated by blue and green boxes. The black lines show untranslated introns. The write boxes show regions. The translation start site (ATG) is indicated by a broken line. Both fosaa gene and fosab gene have 4 exons and the target sites are located in exon 2 by black arrowhead. The black and red letters respectively represent the target region and protospacer adjacent motif (PAM). (B): Mutation pattern of fosaa and fosab knockout. The black and red boxes represent sgRNA target binding site and PAM respectively. The red letters and hyphens represent nucleotide insertion and deletion. (C): The fosaa and fosab deletions and insertions caused frame-shift early termination of translation, resulting in fosaa−/− and fosab−/− proteins with only 70 and 63 amino acids (aa) individually. (D): Left and right panels show PCR genotyping of fosaa and fosab knockout zebrafish. WT, +/− and −/− indicate wild-type, heterozygous and homozygous zebrafish, respectively. (E): Relative mRNA expression of fosaa and fosab in WT zebrafish, fosaa−/− and fosab−/− zebrafish. Data represent mean ± SEM (n = 6). Values marked with * is significantly different with the WT group (P < 0.05) and *** is significantly different with the WT group (P < 0.001).
|

