Fig. 1
- ID
- ZDB-FIG-220509-33
- Publication
- Dulla et al., 2021 - Antisense oligonucleotide-based treatment of retinitis pigmentosa caused by USH2A exon 13 mutations
- Other Figures
- All Figure Page
- Back to All Figure Page
|
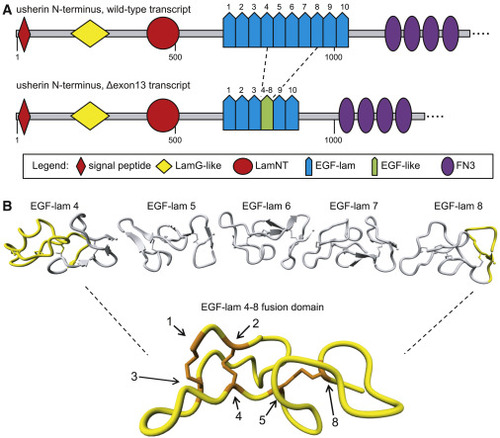
In silico modeling of usherin after exon 13 skipping (A) Schematic representation of the domain architecture of wild-type (WT) usherin and usherinΔexon 13. Individual EGF-lam domains are numbered. Skipping of exon 13 results in the exclusion of EGF-lam domains 5, 6, and 7, as well as the partial exclusion of EGF-lam domains 4 and 8. The remaining amino acids of EGF-lam domains 4 and 8 are predicted to form an EGF-like domain with six cysteine residues. The fusion site of this domain is located between the fifth cysteine residue of EGF-lam domain 4 and the eighth cysteine derived from EGF-lam 8. (B) 3D homology modeling predicts the formation of a stable EGF-like domain with normal disulfide bridge formations. The predicted structure of usherin EGF-lam domains 4 (left) and 8 (right) are shown. The amino acids that are encoded by USH2A exon 13 are depicted in gray and predicted to be absent after translation of USH2A Δexon 13 transcripts. Cysteine residues that are present in the EGF-like fusion domain are numbered and indicated in orange. The cysteine residues numbered 1 to 5 are derived from EGF-lam domain 4, whereas residue 8 is derived from EGF-lam domain 8. |