- Title
-
Antisense oligonucleotide-based treatment of retinitis pigmentosa caused by USH2A exon 13 mutations
- Authors
- Dulla, K., Slijkerman, R., van Diepen, H.C., Albert, S., Dona, M., Beumer, W., Turunen, J.J., Chan, H.L., Schulkens, I.A., Vorthoren, L., Besten, C.D., Buil, L., Schmidt, I., Miao, J., Venselaar, H., Zang, J., Neuhauss, S.C.F., Peters, T., Broekman, S., Pennings, R., Kremer, H., Platenburg, G., Adamson, P., de Vrieze, E., van Wijk, E.
- Source
- Full text @ Mol. Ther.
|
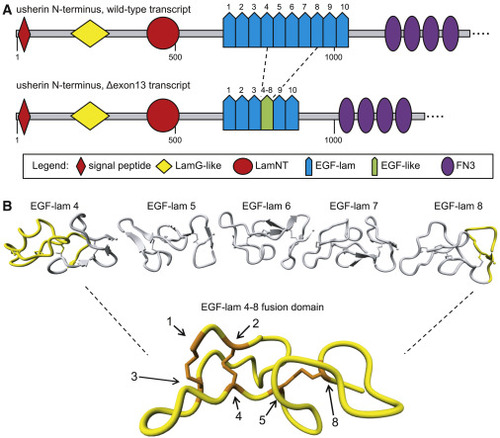
In silico modeling of usherin after exon 13 skipping (A) Schematic representation of the domain architecture of wild-type (WT) usherin and usherinΔexon 13. Individual EGF-lam domains are numbered. Skipping of exon 13 results in the exclusion of EGF-lam domains 5, 6, and 7, as well as the partial exclusion of EGF-lam domains 4 and 8. The remaining amino acids of EGF-lam domains 4 and 8 are predicted to form an EGF-like domain with six cysteine residues. The fusion site of this domain is located between the fifth cysteine residue of EGF-lam domain 4 and the eighth cysteine derived from EGF-lam 8. (B) 3D homology modeling predicts the formation of a stable EGF-like domain with normal disulfide bridge formations. The predicted structure of usherin EGF-lam domains 4 (left) and 8 (right) are shown. The amino acids that are encoded by USH2A exon 13 are depicted in gray and predicted to be absent after translation of USH2A Δexon 13 transcripts. Cysteine residues that are present in the EGF-like fusion domain are numbered and indicated in orange. The cysteine residues numbered 1 to 5 are derived from EGF-lam domain 4, whereas residue 8 is derived from EGF-lam domain 8. |
|
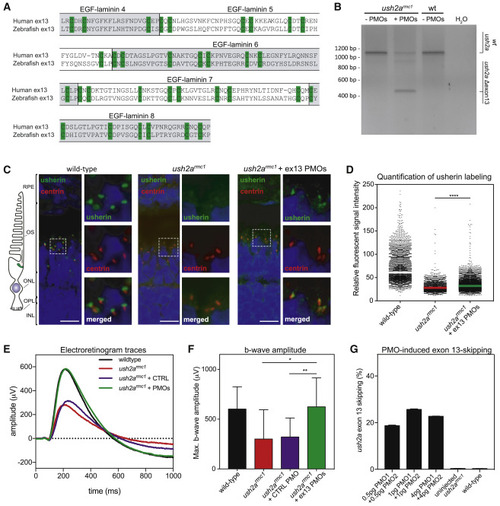
Morpholino antisense oligonucleotides (AONs) mediate ush2a exon 13 skipping, usherinΔexon 13 protein expression, and restoration of electroretinogram (ERG) in a mutant zebrafish model (A) Amino acid alignment of the sequences encoded by human and zebrafish USH2A exon 13. The (partial) EGF-lam domains are indicated. The cysteine residues required for 3D topology of the EGF-lam domains (green) are completely conserved between zebrafish and human. (B) Phosphorodiamidate morpholino oligonucleotide (PMO)-induced skipping of ush2a exon 13 in zebrafish larvae. ush2armc1 mutant embryos were injected with a combination of PMO1 and PMO2 (1 ng of each). Investigation of ush2a pre-mRNA splicing at 3 days post-fertilization (dpf) revealed the skipping of ush2a exon 13 upon injection of PMOs targeting ush2a exon 13. Uninjected ush2armc1 mutant zebrafish larvae and WT larvae were used as controls. (C) Subcellular localization of usherin in horizontal cryosections of larval (5 dpf) zebrafish retinae. Usherin was visualized with anti-usherin antibodies directed against the intracellular C-terminal tail of zebrafish usherin (green signal). Nuclei were stained with DAPI (blue signal), and the connecting cilium is labeled using anti-centrin antibodies (red). In WT larvae, usherin is present at the photoreceptor periciliary membrane, adjacent to the connecting cilium. In homozygous ush2armc1 larvae, no specific usherin signal could be detected. PMO-induced ush2a exon 13 skipping in ush2armc1 mutant larvae resulted in partial restoration of usherinΔexon 13 expression with the correct subcellular localization in the retina. OS, outer segment; ONL, outer nuclear layer; OPL, outer plexiform layer; IPL, inner plexiform layer; wt: WT; ush2armc1, zebrafish with exon 13 mutation. (D) Scatterplot of the relative fluorescence intensity of anti-usherin staining in the periciliary region of all photoreceptors in the middle section of the larval zebrafish eye. The signal intensity is decreased in the ush2armc1 retina compared to WTs. Relative fluorescent signal intensity of anti-usherin staining is significantly increased in PMO-injected ush2armc1 mutants as compared to uninjected mutants (∗∗∗∗p < 0.0001, Kruskal-Wallis test followed by Dunn’s nonparametric post-test). (E) Average ERG b-wave traces from uninjected, control PMO-injected, exon 13 PMO-injected ush2armc1 larvae and WT controls at 5−6 dpf. PMO-induced skipping of ush2a exon 13 completely restored b-wave amplitudes in ush2armc1 larvae as compared to uninjected or control PMO-injected mutants. (F) Maximum b-wave amplitudes recorded in uninjected or control PMO-injected ush2armc1 larvae are significantly reduced as compared to ERG traces from age- and strain-matched WT controls. Maximum b-wave amplitudes recorded in PMO-injected ush2armc1 mutants are significantly improved as compared to ERG traces from uninjected or control PMO-injected ush2armc1 mutants and do not significantly differ from WTs (p > 0.99). Data are shown as mean ± SD, ∗p < 0.05, ∗∗p < 0.01, Kruskal-Wallis test followed by Dunn’s nonparametric post-test. (G) Quantification of ush2a Δexon 13 transcripts in uninjected and PMO-injected zebrafish larvae at 3 dpf. EXPRESSION / LABELING:
|
|
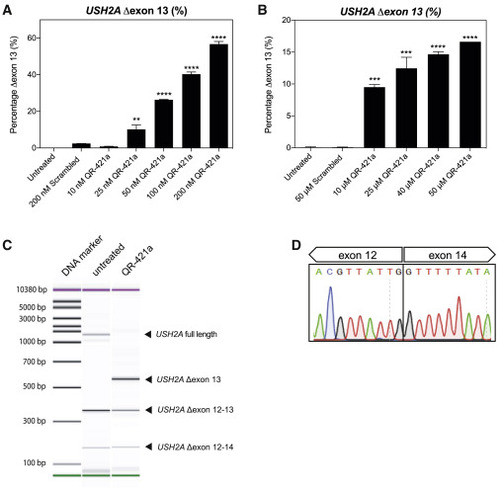
QR-421a shows a concentration-dependent increase of USH2A exon 13 skipping in WERI-Rb1 cells (A and B) WERI-Rb1 retinoblastoma cells were treated with different concentrations of QR-421a, using either (A) transfection or (B) gymnotic uptake. Untreated and control oligo-treated cells were included as negative controls. Exon-skipping level was determined by quantification of USH2A transcripts with and without exon 13 by RT-ddPCR. Treatment with QR-421a resulted in a significant concentration-dependent increase of USH2A Δexon 13 transcripts. Data are shown as mean ± SD. Two biological replicates per treatment condition. Asterisks indicate significant differences with scrambled control oligo-treated cells (∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; one-way ANOVA followed by Dunnett’s multiple comparison test). (C) Representative image of exon 11-15 RT-PCR amplicons obtained with RNA isolated from untransfected and QR-421a-transfected WERI-Rb1 cells. QR-421a is able to induce skipping of USH2A exon 13 and does not increase the formation of other alternatively spliced USH2A transcripts. Of note, USH2A Δexon 12-13 and Δexon 12-14 transcripts are already present in untreated WERI-Rb1 cells and yield out-of-frame mRNA transcripts. (D) Sanger sequencing traces of the USH2A Δexon 13 amplicon shown in (C) confirm that the sequence of exon 13 is lacking from the transcript. |
|
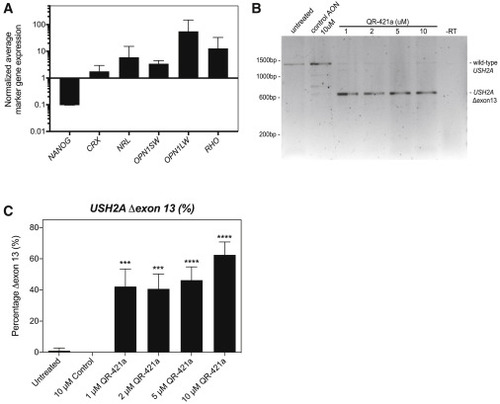
QR-421a treatment induces a concentration-dependent increase of USH2A exon 13 skipping in iPSC-derived photoreceptor progenitor cells (PPCs) from a patient (USH2Ac.2299delG/c.2299delG) (A) Gene-expression analysis indicates successful differentiation toward PPCs. The decrease in NANOG expression is indicative for loss of pluripotency, whereas the increased expression of photoreceptor markers CRX, NRL, OPN1SW, OPN1LW, and RHO is indicative of the successful differentiation toward photoreceptor cells. (B) RT-PCR analysis of USH2A exons 11 to 15 in untreated PPCs of a patient homozygous for the c.2299delG mutation only revealed an amplicon containing exons 11 to 15. Continuous treatment of PPCs with QR-421a (28 days) specifically induced the skipping of USH2A exon 13 from the transcript. There was no evidence of alternative splice site activation in exon 13 or skipping of multiple exons. (C) Quantitative analysis of USH2A exon 13 skipping by RT-ddPCR upon continuous treatment with QR-421a. Treatment was started after 3 months of differentiation and lasted for 28 days. One-half of the culture medium was refreshed every other day with medium containing QR-421a. Skipping of exon 13 was already observed at the lowest concentration and further increased with increasing concentrations. Asterisks indicate significant differences with scrambled control oligo-treated cells (∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; mean ± SD of 3 samples per condition, one-way ANOVA followed by Dunnett’s multiple comparison test). |
|
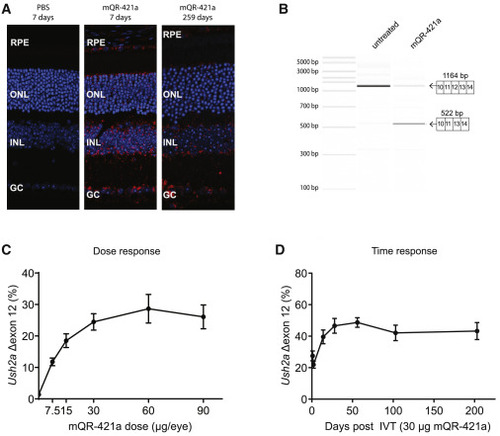
Retinal uptake, efficacy, and duration of action of mQR-421a upon intravitreal (IVT) injection in WT mice (A) Representative sections of untreated and mQR-421a-treated mouse retina. WT C57BL/6J mice received a single bilateral IVT injection of 50 μg of mQR-421a and were sacrificed to visualize the presence of mQR-421a at 7 and 259 days post-injection. mQR-421a was visualized using a Cy5-labeled FISH assay (red signal); cell nuclei were stained with DAPI (blue signal). GC, ganglion cell layer; INL, inner nuclear layer; RPE, retinal pigment epithelium. (B) Bioanalyzer (PCR) analysis of Ush2a exon 12 skipping in untreated and mQR-421a-treated mice. Ush2a transcripts were amplified using primers binding to exons 10 and 14. The top 1,164-base pair band corresponds to Ush2a transcripts with exon 12, and the bottom 522-base pair band corresponds to transcripts without exon 12. (C) Dose-dependent Ush2a exon 12 skipping by mQR-421a in WT C57BL/6J mice. Mice received a single bilateral IVT of 7.5, 15, 60, or 90 μg per eye of mQR-421a or 30 μg control AON. Ush2a transcripts with or without exon 12 were quantified by ddPCR and normalized by total Ush2a levels measured at exon 8-9, and percentage of exon 12 skipping was calculated. Mean ± SEM, n = 6 animals per dose per group. (D) Long duration of action of mQR-421a in mice retina. Mice received a single bilateral IVT of 30 μg mQR-421a per eye and followed for 1, 2, 14, 28, 56, 103, or 203 days. Ush2a transcripts with or without exon 12 were quantified by ddPCR and normalized by total Ush2a levels measured at exon 8-9, and percentage of exon 12 skipping was calculated. Mean ± SEM, n = 12 (1, 2, 14, and 28 day time points) or n = 8 (56, 103, and 203 day time point) eyes. |