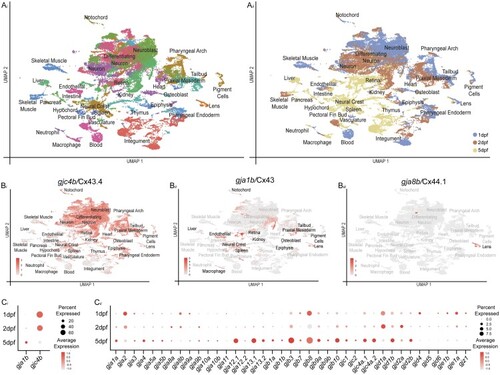
Fig. 1.
- ID
- ZDB-FIG-220509-1
- Publication
- Lukowicz-Bedford et al., 2022 - Connexinplexity: The spatial and temporal expression of connexin genes during vertebrate organogenesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
scRNA-seq dataset of zebrafish organogenesis and |
| Gene: | |
|---|---|
| Fish: | |
| Anatomical Term: | |
| Stage: | Prim-5 |