- Title
-
Connexinplexity: The spatial and temporal expression of connexin genes during vertebrate organogenesis
- Authors
- Lukowicz-Bedford, R.M., Farnsworth, D.R., Miller, A.C.
- Source
- Full text @ G3 (Bethesda)
|
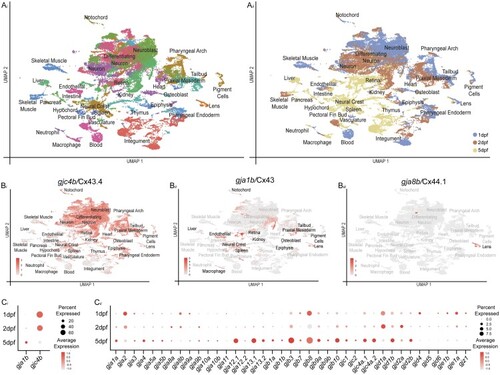
scRNA-seq dataset of zebrafish organogenesis and EXPRESSION / LABELING:
|
|
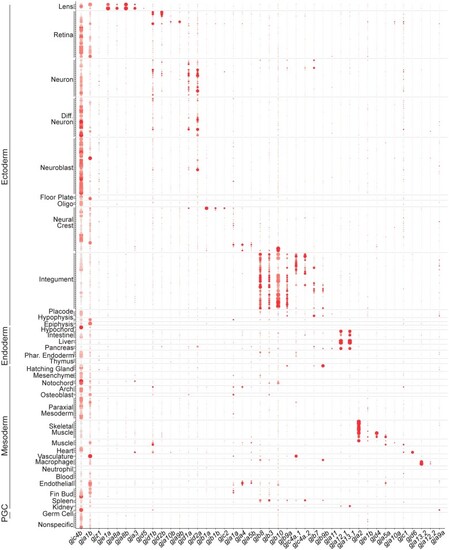
Connexin expression during zebrafish organogenesis. Clusters are organized by annotations and grouped into tissues and germ layers denoted on the |
|
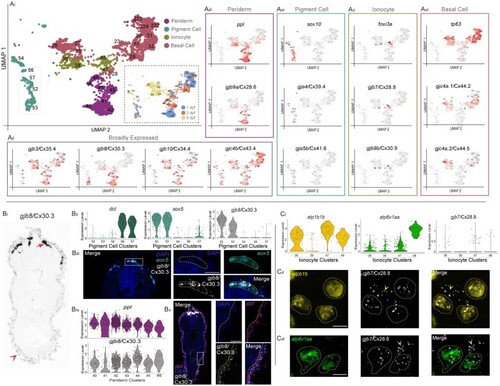
Connexin expression in the zebrafish integument during organogenesis. a EXPRESSION / LABELING:
|