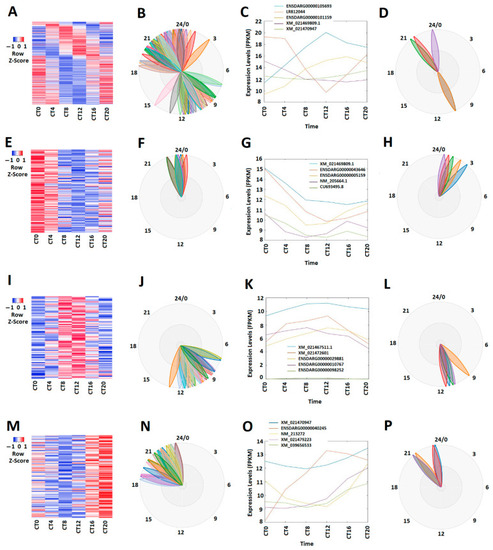
Figure 1
|
Expression profile analysis of circadianly expressed zebrafish larval lncRNAs under the DD condition. (A–D) Analysis of all the 269 circadianly expressed larval lncRNAs under the DD condition. Heat map (A) and BioDare2 plots (B) of all the 269 circadianly expressed zebrafish larval lncRNAs, expression profiles (C) and BioDare2 plots (D) of representative lncRNAs. (E–H) Analysis of 100 larval morning (CT0 and CT4) lncRNAs. Heat map (E) and BioDare2 plots (F) of the 100 larval zebrafish morning lncRNAs, expression profiles (G) and BioDare2 plots (H) of zebrafish larval morning representative lncRNAs. (I–L) Analysis of 75 zebrafish larval evening (CT8 and CT12) lncRNAs. Heat map (I) and BioDare2 plots (J) of the 75 larval evening lncRNAs, expression profiles (K) and BioDare2 plots (L) of larval evening representative lncRNAs. (M–P) Analysis of 94 larval zebrafish night (CT16 and CT20) lncRNAs. Heat map (M) and BioDare2 plots (N) of the 94 larval zebrafish night lncRNAs, expression profiles (O) and BioDare2 plots (P) of larval zebrafish night representative lncRNAs. |