FIGURE 4
- ID
- ZDB-FIG-211029-77
- Publication
- Gans et al., 2021 - Glucocorticoid-Responsive Transcription Factor Krüppel-Like Factor 9 Regulates fkbp5 and Metabolism
- Other Figures
- All Figure Page
- Back to All Figure Page
|
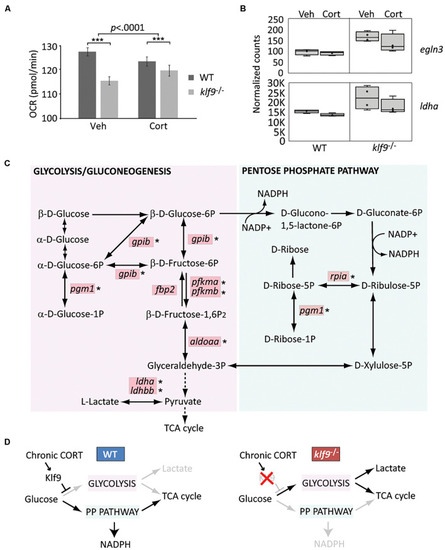
Klf9 regulates metabolism. (A) Oxygen consumption rate (OCR) at 1 dpf, as measured by Seahorse XF96 extracellular flux analyzer. The averages of three experimental replicates are shown, each with 20 embryos per condition measured individually and simultaneously over the course of at least 85 min (thus n = 60 total for each condition). The bars represent the average Y-intercept of linear fits of the averaged time-course data (see Supplementary Figure 12), ±standard error. Two-way ANOVA indicated significant effects of both genotype (WT vs. klf9–/–, p < 0.0001) and treatment (VEH vs. CORT, p = 0.04), as well as a significant interaction (p < 0.0001). ***Adjusted p < 0.0001 by Šídák’s multiple comparisons test. (B) Normalized counts of Hif1a target egln3 and glycolytic gene ldha (from Gans et al., 2020). (C) Krüppel-like factor 9 (Klf9) downregulates multiple genes in the glycolysis/gluconeogenesis pathway. Red boxes indicate genes significantly over expressed (FDR < 0.05) in RNA-seq of klf9–/– larvae compared to wild type (Gans et al., 2020). Asterisks denote genes in which putative Klf9 binding sites were identified using HOMER. (D) Model for Klf9-mediated regulation of metabolic flux. In response to stress, Klf9 is predicted to inhibit glycolysis, shunting flux through the Pentose Phosphate Pathway (PPP). |
| Fish: | |
|---|---|
| Condition: | |
| Observed In: | |
| Stage: | Prim-5 |