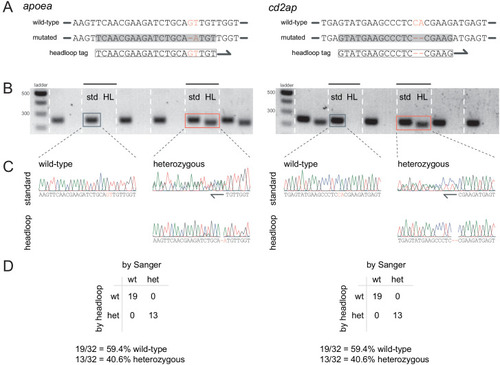
Figure 3—figure supplement 2.
- ID
- ZDB-FIG-210113-60
- Publication
- Kroll et al., 2021 - A simple and effective F0 knockout method for rapid screening of behaviour and other complex phenotypes
- Other Figures
- All Figure Page
- Back to All Figure Page
|
( |