Fig. 1
- ID
- ZDB-FIG-201209-36
- Publication
- Zhao et al., 2020 - An optimized base editor with efficient C-to-T base editing in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
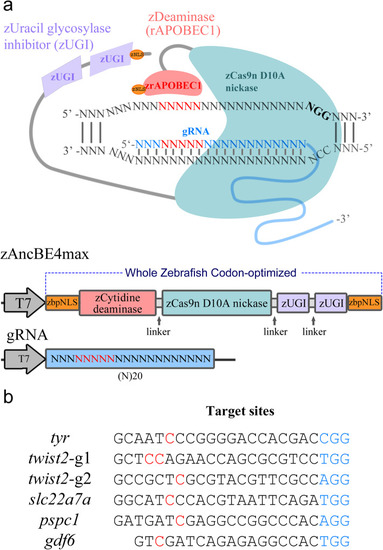
Base editor in zebrafish using a whole zebrafish codon-optimized BE4 system. |