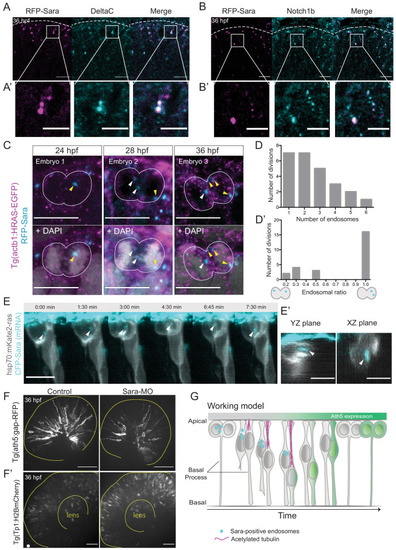
Figure 5—figure supplement 1.
- ID
- ZDB-FIG-201125-14
- Publication
- Nerli et al., 2020 - Asymmetric neurogenic commitment of retinal progenitors involves Notch through the endocytic pathway
- Other Figures
-
- Figure 1—figure supplement 1.
- Figure 1—figure supplement 1.
- Figure 2—figure supplement 1—source data 1.
- Figure 2—figure supplement 1—source data 1.
- Figure 3—figure supplement 1.
- Figure 3—figure supplement 1.
- Figure 3—figure supplement 2.
- Figure 4—video 1.
- Figure 5—figure supplement 1.
- Figure 5—figure supplement 1.
- All Figure Page
- Back to All Figure Page
|
( |