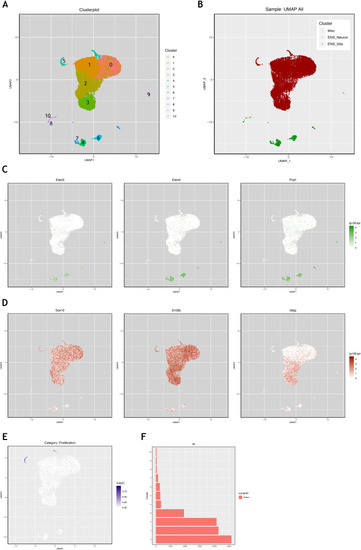
Figure 2—figure supplement 3.
- ID
- ZDB-FIG-201003-96
- Publication
- McCallum et al., 2020 - Enteric glia as a source of neural progenitors in adult zebrafish
- Other Figures
-
- Figure 1
- Figure 1—figure supplement 1.
- Figure 2
- Figure 2—figure supplement 1.
- Figure 2—figure supplement 2.
- Figure 2—figure supplement 3.
- Figure 3
- Figure 3—figure supplement 1.
- Figure 4
- Figure 4—figure supplement 1.
- Figure 5
- Figure 5—figure supplement 1.
- Figure 6
- Figure 6—figure supplement 1.
- Figure 6—figure supplement 2.
- Figure 6—figure supplement 3.
- Figure 7
- Figure 7—figure supplement 1.
- All Figure Page
- Back to All Figure Page
|
Using the mouse single cell transcriptomic data from |