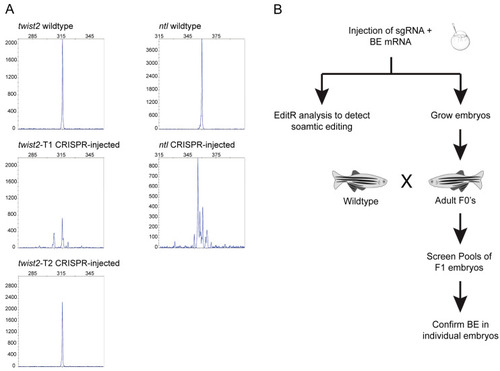
Figure 2
- ID
- ZDB-FIG-200814-18
- Publication
- Carrington et al., 2020 - BE4max and AncBE4max Are Efficient in Germline Conversion of C:G to T:A Base Pairs in Zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
Selection of targets and strategy for assessment of base editing efficiency. ( |