Fig. 5
- ID
- ZDB-FIG-200528-3
- Publication
- Tamaoki et al., 2020 - Splicing- and demethylase-independent functions of LSD1 in zebrafish primitive hematopoiesis
- Other Figures
- All Figure Page
- Back to All Figure Page
|
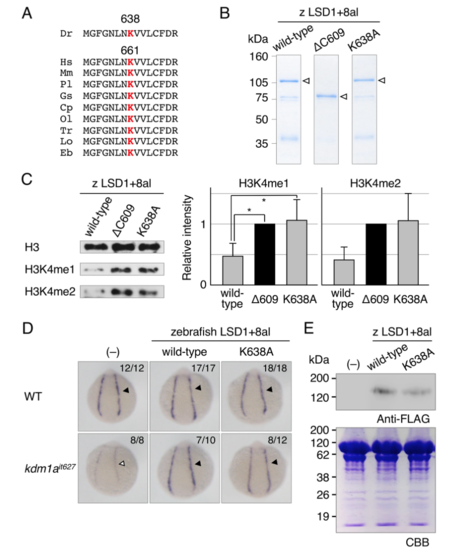
Phenotypic rescue experiments using demethylase-deficient zebrafish LSD1. (A) Multiple alignments of vertebrate LSD1 proteins around Lys661 in human LSD1. Red “K” indicates highly conserved lysine residues in the catalytic center of LSD1. (B) Bacterially expressed zebrafish LSD1+8al proteins used in a demethylation assay. Arrowheads indicate full-length proteins. (C) A demethylation assay of indicated zebrafish LSD1+8al proteins using bulk histones from calf thymus as substrates. Methylated proteins were detected by immunoblotting using specific antibodies. Bar graphs indicate the relative intensity of immunoblot signals standardized to the level of a negative control LSD1+8alΔ609. Error bars represent standard deviations. The experiments was repeated 3 times. Asterisks denote p values < 0.05. (D) The gata1a expression at 15 hpf in the lateral plate mesoderm (arrowheads) of wild-type or kdm1ait627 embryos injected with or without mRNAs encoding wild-type or K638A mutant-type zebrafish LSD1+8al. (E) Immunoblot analysis using anti-FLAG antibody of FLAG-tagged LSD1 proteins in 15-hpf wild-type embryos. Total protein loading was shown by CBB staining. |