Fig. 4
- ID
- ZDB-FIG-200406-284
- Publication
- Ohata et al., 2020 - An Activity-Based Methionine Bioconjugation Approach To Developing Proximity-Activated Imaging Reporters
- Other Figures
- All Figure Page
- Back to All Figure Page
|
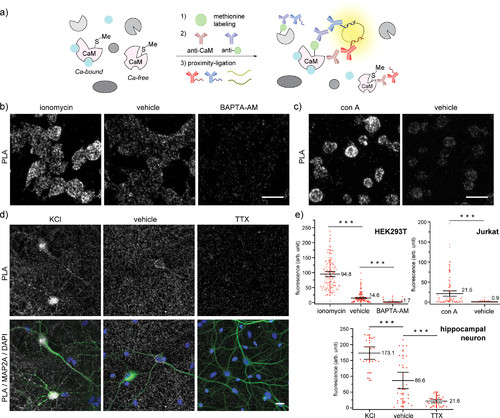
“Turn-on” detection of changes in calcium levels in cells by applying the methionine proximity-activated imaging reporter (Met-PAIR) method to the native calcium regulatory protein calmodulin (CaM) at endogenous levels. Oxaziridine labeling in live cells was conducted in the presence of various reagents in Hanks’ balanced salt solution (HBSS) with alkyne-tagged oxaziridine (Ox4, 20 μM) at rt for 20 min (HEK293T and Jurkat) or with Ox4 (0.5 μM) at 37 °C for 5 min (hippocampal neurons). Proximity-ligation assay (PLA) staining was performed with mouse anti-CaM antibody and rabbit anti-Oregon Green antibody (HEK293T and Jurkat) or rabbit anti-biotin antibody (neuron). Gray: PLA staining. Blue: DAPI nuclear staining. Scale bar: 20 μm. (a) A schematic illustration of Met-PAIR on CaM. Blue ball: calcium ions in live cells. (b) Confocal images of HEK293T cells stained by Met-PAIR with intracellular calcium increases (ionomycin: 10 μM) or decreases (BAPTA-AM: 30 μM). (c) Confocal images of Jurkat cells stained by Met-PAIR with concanavalin A (con A, 0.1 mg/mL) to trigger calcium increases over vehicle control. (d) Confocal images of rat hippocampal neuron stained by Met-PAIR with activation (KCl: 90 mM) or deactivation (tetrodotoxin, TTX: 10 μM) to increase or decrease calcium transients, respectively. Anti-MAP2A antibody (green) was used to identify neurons. (e) Whisker plots for the confocal images in (b) HEK293T, (c) Jurkat, and (d) neurons. Each dot represents the fluorescence intensity of single cells or neuronal cell bodies. Whisker and center line represent 95% confidence interval and mean intensity, respectively. The mean value is shown near the center line. The quantification was conducted by imaging 4 regions in each of 2 independent biological replicates for HEK293T and Jurkat (total 8 cell images), and 5 regions in each of 2 independent biological replicates (total 10 images) were used for the quantification of neuronal cell bodies identified by the MAP2A staining. The number of quantified cells: 139 (ionomycin), 164 (HEK293T vehicle), 170 (BAPTA-AM), 113 (con A), 105 (Jurkat vehicle), 24 (KCl), 30 (neuron vehicle), and 32 (TTX). ***P < 0.001, Student’s t test. |