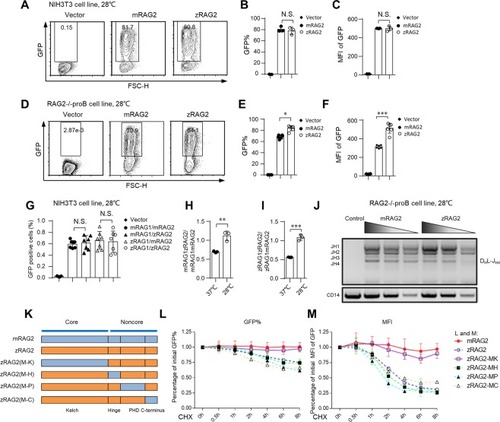
The protein stability and recombination efficiency of zRAG2 are similar to those of mRAG2 at 28 °C. (A–F) The zRAG2 protein showed an expression level similar to that of the mRAG2 protein, and its MFI was also similar to that of mRAG2 or even higher at 28 °C. zRAG2-GFP fusion protein and mRAG2-GFP fusion protein were introduced into NIH3T3 cells (A–C) or RAG2-deficient pro-B cells (D–F) by the retrovirus-mediated gene transfer method. The cells were first incubated for 36 h at 37 °C, and they were transferred to 28 °C after this time. After 48 h at 28 °C, GFP expression and intensity were analyzed by FACS. (G) NIH3T3 cells were transfected with pCJGFP using Lipo6000 transfection reagent. The GFP level was measured by flow cytometry to assess the recombination efficiency. The percentages of GFP-positive cells were shown (the means ± SDs were calculated from triplicate experiments). (H,I) The RAG1zRAG2/mRAG1mRAG2 and zRAG1zRAG2/zRAG1mRAG2 ratios were shown. (J) PCR analyses of the indicated DHJH family rearrangements in RAG2-deficient pro-B cells at 28 °C. Input control: CD14 (bottom). PCR amplification was performed with fivefold serial dilutions of genomic DNA. The results are typical of three experiments. Bands related to rearrangements of various JH segments are indicated on the left. (K–M) Different mouse RAG2 domains were substituted for the corresponding zRAG2 domains. GFP% (L) and MFI (M) are shown from 0 h to 8 h after treatment with the 20 μg/ml CHX. MK, mouse Kelch; MH, mouse hinge; MP, mouse PHD; MC, mouse C-terminus. The error bars indicate the SDs. The data are presented as the mean ± standard deviation. *P < 0.05, **P < 0.01 and ***P < 0.001 by Student’s t test; N.S.: no significance.
|

