|
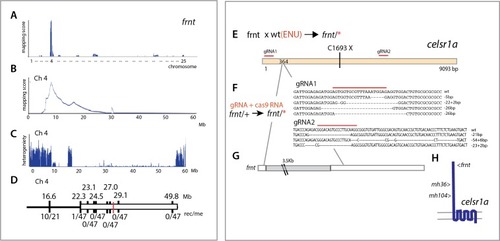
Identification of altered <italic>celsr1a</italic> function underlying the <italic>frnt</italic> phenotype.(A) Mapping by homozygosity-by-descent indicates linkage of frnt to chromosome 4. (B) Mapping score across chromosome 4. (C) Analysis of heterogeneity across chromosome four showing a broad region of homogeneity in the population indicating linkage. (D) Fine mapping of frnt showing limited recombination and resolution of the map position along chromosome 4; white bar, area showing linkage; red hashmark, position of celsr1a; top, position (megabase, Mb) on chromosome 4, zv9 assembly (https://ensembl.org); bottom number of recombinants per meiosis (rec/me) scored. (E) Chemical mutagenesis loss-of-complementation screen to identify the gene mutation underlying the frnt phenotype. Exome sequencing of identified founders having the frnt phenotype (frnt/*), identified mutations in the celsr1a gene within the mapped interval (mh36, C1693X). (F) Identified deletions/insertions within celsr1a generated through CRISPR/Cas9 genome editing that fail to complement frnt. Recovered sequences from F1 founders; guideRNA position demarcated with overlain red bar. Of the recovered lines, allele mh104_P2027A-fs11X was retained G) Identification of transposon insertion within celsr1a in frnt.H) Schematic of celsr1a and position of identified mutations.
|