Fig. S2
- ID
- ZDB-FIG-200116-42
- Publication
- Frikstad et al., 2019 - A CEP104-CSPP1 Complex Is Required for Formation of Primary Cilia Competent in Hedgehog Signaling
- Other Figures
- All Figure Page
- Back to All Figure Page
|
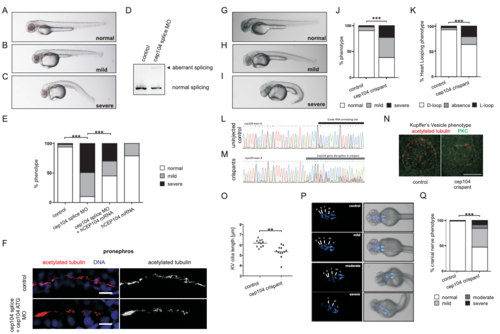
Phenotype of zebrafish treated with a cep104 splice blocking morpholino and ciliopathy phenotypes in cep104 zebrafish crispants (related to Figure 1) (A‐C) 48 hpf zebrafish display pericardial edema and cardiac defects following cep104 splice MO injection, with varying severity from mild (B) to severe (C). (D) Reverse‐transcriptase PCR indicating incorrect splicing (upper band, arrow head) of the cep104 transcript in cep104 splice MO-injected embryos. (E) 48 hpf zebrafish were scored under light microscopy for phenotypes (normal/mild/ severe). Phenotypic rescue of cep104 splice MOinjected embryos with co‐injection of human CEP104 mRNA. Chi square test of independence revealed that the frequencies of phenotypes are significantly different between uninjected controls and morphants (***p<0.0001) and between morphants and morphants co-injected with CEP104 mRNA (***p<0.0001; Control n= 100; cep104 splicing MO n= 150; cep104 splicing MO + CEP104 mRNA n= 160; CEP104 mRNA n=76). (F) IFM of cilia in distal pronephros in control and cep104 morphant embryos at 72 hpf does not reveal an obvious ciliary phenotype in pronephros of cep104 morphant fish. a-acetylated tubulin, red (left panels), white (right panels), DAPI, blue. Scale bars = 10μm. (G-I) Injection of Cas9 protein along with specific guide RNA targeting intron 7/exon 8 of zebrafish cep104 gene results in crispant embryos that phenocopy cep104 morphant embryos, with varying severity from mild (H) to severe (I) when compared to uninjected control (G) at 48 hpf. Crispants exhibited microophthalmia, with a quantified reduction in area expressed as a ratio to control embryos of 0.81, p=0.0038 (unpaired t-test, n = 18 vs. 7 control). Severe crispants and morphants showed some yolk sac abnormalities, which are likely to be linked to pericardial edema. (J) Percentage of control and crispant zebrafish displaying phenotypes. (*** p<0.0001, Chi square test, control n= 229; cep104 crispants n= 240). (K) cep104 crispants display heart looping defects at 48 hpf. Percentage control and crispant fish displaying phenotypes is shown. (*** p<0.0001, Chi square test, control n= 209; cep104 crispants n= 155). (L-M) Sequence chromatograms showing genomic cep104 sequence in an uninjected control embryo (L) and in an embryo injected (M) with Cas9 protein along with specific guide RNA (black bar) targeting cep104. Injection results in genetic mosaic crispant embryos (F0) carrying various mutations in the cep104 gene (M). Mutations in cep104 gene were found in 91% of embryos screened (n= 11). (N,O) IFM of cilia (anti-acetylated tubulin, red) in Kupffer’s Vesicle (KV) at the 10-somite stage in control and cep104 crispant embryos reveals a reduction in cilia length. Cell junctions are stained with anti-PKC (green). Scale bar 50 μm. (O) Dot plots confirming a significant reduction in the length of cilia present in KV in cep104 crispant zebrafish (n=11) when compared to uninjected control (n=12) (** p<0.001, unpaired Student’s t-test). (P) Dorsal views of 48 hpf islet-1:GFP transgenic fish imaged under fluorescence and light microscopy. Injection with Cas9 protein along with sgRNA leads to cranial nerves defects, with varying severity from mild to moderate and severe when compared to uninjected control. (Q) Percentage control and crispant fish displaying phenotypes is shown. (*** p<0.0001, Chi square test, control n= 106; cep104 crispant n= 157).
|
| Fish: | |
|---|---|
| Knockdown Reagents: | |
| Observed In: | |
| Stage Range: | 10-13 somites to Protruding-mouth |