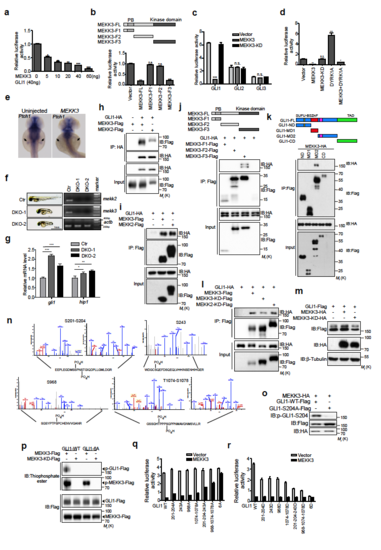
MEKK2/3 specifically inhibit GLI1 transcriptional activity, associate with and phosphorylate GLI1 (a) MEKK3 inhibited GLI1 transcriptional activity in a dose-dependent manner. HEK293T cells were co-transfected with GliBS-luc reporter, GLI1 and MEKK3. Cell lysates were analyzed using a luciferase assay to measure GLI1 transcriptional activity. (b) The kinase domain of MEKK3 inhibited GLI1 transcriptional activity. A diagram shows the truncated mutants of MEKK3 (Upper panel). HEK293T cells were co-transfected with GliBS-luc reporter, GLI1, and MEKK3 truncated mutants. Cell lysates were analyzed by luciferase assay. (c) MEKK3 did not inhibit GLI2 and GLI3 transcriptional activity using GliBS-luc reporter assay in HEK293T cells. (d). MEKK3 inhibited DYRK1A induced Gli1 transcriptional activity. HEK293T cells were co-transfected with GliBS-luc reporter, GLI1 and indicated plasmids. Cell lysates were analyzed using a luciferase assay. (e) Whole-mount in situ hybridization of ptch1 in uninjected zebrafish embryos and embryos injected with human MEKK3 mRNA. Embryos were collected at 42 h post-fertilization (hpf). ptch1 expression in the fin buds indicated by arrows was significantly reduced in embryos injected with MEKK3 mRNA (n=35/47). (f) The gross appearance of the DKO zebrafish embryos for mekk2 and mekk3. RT-PCR analysis revealed the lack of expression of mekk2 and mekk3 in DKO zebrafish embryos. (g) qPCR analysis of gli1 and hip1 expression in control and DKO zebrafish exbryos. (h) and (i) Co-immunoprecipitation assay revealed that GLI1 forms complexes with MEKK2 and MEKK3. Lysates from HEK293T cells transfected with indicated plasmids were immunoprecipitated and immunoblotted as indicated. (j) Kinase domain of MEKK3 associated with GLI1 in a co-immunoprecipitation assay using a series of MEKK3 deletion mutants in HEK293T cells. (k) A series of GLI1 deletion mutants were constructed as indicated and transfected into HEK293T cells together with MEKK3-HA. Cell lysates were immunoprecipitated with anti-Flag antibody and then subjected to western blot analysis using indicated antibodies. SUFU-BS, SUFU binding site; ZnF, zinc finger domain; NLS, nuclear localization signal; NES; nuclear export signal; TAD, transcriptional-activation domain. (l) GLI1 associated with kinase dead forms of MEKK2 and MEKK3 in a co-immunoprecipitation assay in HEK293T cells. (m) MEKK3 induced a mobility shift of exogenous GLI1 in HEK293T cells. (n) Mass spectrometric analysis of GLI1 phosphorylation by MEKK3. HEK293T cells were co-transfected with GLI1-Flag and MEKK3-HA plasmids. The lysates were subjected to immunoprecipitation using anti Flag antibody, and GLI1 protein band was isolated and subjected to mass spectrometry. Extracted ion chromatograms (EICs) identified phosphorylated peptides at S201, S204, S243, S968, T1074 and S1078 in GLI1. (o) p-GLI1-S204 antibody recognized WT GLI1-Flag but not GLI1-S204A-Flag co-expressed with MEKK3 in HEK293T cells, indicating that this antibody is highly specific. (p) GLI1-6A mutant was resistant to phosphorylation by MEKK3 in an in vitro kinase assay. (q) and (r) GliBS-luc reporter assay evaluated the contribution of different phosphorylation sites to the GLI1 transcriptional activity in HEK293T cells. *P < 0.05, **P < 0.01 and ***P < 0.001 (two-tailed Student’s t-test). All data were mean ± s.d. from representative of three independent experiments conducted in triplicate.
|

