Fig. 5
|
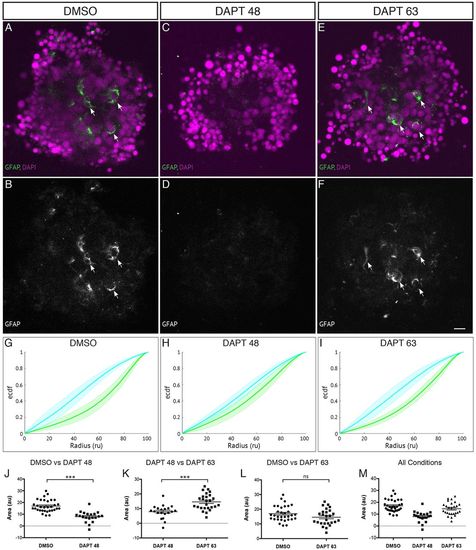
Late application of DAPT allows Müller glia to be generated and aggregates to self-organise. Aggregates are cultured in the presence of DAPT applied at 45-48 hpf onwards to block the differentiation of Müller glia or at 63 hpf onwards to allow the differentiation of some Müller glia compared with DMSO control. (A,B) Aggregates cultured in the presence of DMSO show several GFAP-positive cells (indicated by arrows). (C,D) Aggregates cultured in the presence of DAPT from 45-48 hpf onwards have little or no GFAP-positive cells. (E,F) Aggregates cultured in the presence of DAPT from 63 hpf onwards have several GFAP-positive cells (indicated by arrows). Scale bar: 10μm. (G) Average ECDF plots for aggregates treated with DMSO. (H) Average ECDF plots for aggregates treated with DAPT from 45-48 hpf onwards. (I) Average ECDF plots for aggregates treated with DAPT from 63 hpf onwards. (J) Area (in arbitrary units) is calculated between the ECDF for the Crx:gapCFP population and the ECDF for the Ptf1a:cytGFP population of cells, and compared between aggregates treated with DMSO and aggregates treated with DAPT from 45-48 hpf onwards (n=32 for DMSO, n=20 for DAPT at 45-48 hpf, Mann–Whitney two-tailed t-test, P<0.0001). (K) Area (arbitrary units) compared between the ECDF plots of aggregates treated with DAPT from 45-48 hpf onwards and aggregates treated with DAPT from 63 hpf onwards (n=20 for DAPT at 45-48 hpf, n=26 for DAPT at 63 hpf, Mann–Whitney two-tailed t-test, P<0.0001). (L) Area (arbitrary units) compared between the ECDF plots of aggregates treated with DMSO and aggregates treated with DAPT from 63 hpf onwards (n=32 for DMSO, n=26 for DAPT at 63 hpf, Mann–Whitney two-tailed t-test, P>0.05). (M) Areas of data from all conditions. |