|
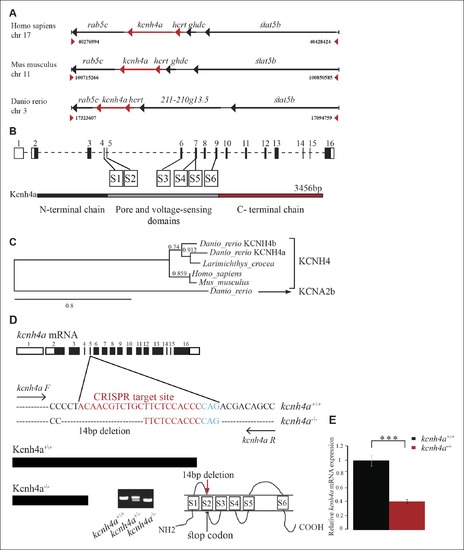
The genomic location, phylogenetic reconstruction and structure of kcnh4a, and the generation of kcnh4a-/- zebrafish. (A) Synteny analysis shows similar genomic context of hcrt in zebrafish and mammals. Notably, kcnh4a is located a few kbs downstream to hcrt in zebrafish and mammals. (B) The 16-exon kcnh4a gene (black box = exon, white box = UTR) encodes for a voltage-gated potassium channel that includes an N-terminal chain (black bar), pore and voltage-sensing domains (S1-6, grey bar), and the C-terminal chain (red bar). (C) A cladogram-style phylogenetic tree depicting the evolutionary conservation of Kcnh4a protein among vertebrates. The tree shows topography as well as distance indicated by the branch support values above corresponding branches. (D) Generation of CRISPR-mediated kcnh4a-/- zebrafish. A 14 bp deletion was introduced in exon 5 that encodes to the S2 domain. A short mutant allele was visible on agarose gel. (E) Quantitative reverse transcription PCR shows reduction of 59% in the expression levels of kcnh4a mRNA in kcnh4a-/- 6 dpf larvae (p < 0.001).
|