Fig. 5
- ID
- ZDB-FIG-140417-24
- Publication
- Bhatia et al., 2014 - A survey of ancient conserved non-coding elements in the PAX6 locus reveals a landscape of interdigitated cis-regulatory archipelagos
- Other Figures
- All Figure Page
- Back to All Figure Page
|
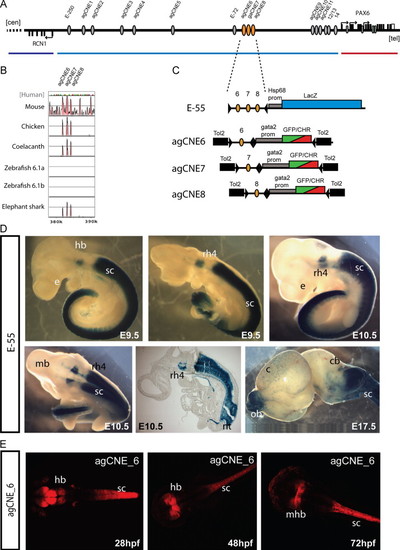
Functional dissection of the E-55 region. (A) Schematic overview of the RCN1 to PAX6 intergenic region with the positions of agCNEs indicated by grey ellipses and agCNE6-8 in orange. (B) VISTA plot of the multispecies sequence alignment of the fragment showing the three peaks of conservation in elephant shark but not in either of the zebrafish pax6 loci. (C) Schematic depiction of the reporter constructs used to generate transgenic mice and fish. (D) Transgenic embryos for the E-55 fragment, containing all three agCNEs, show X-gal staining in rhombomere 4 (rh4) of the hindbrain (hb) and from rhombomere 7 towards caudal along the length of the spinal cord, covering its full area on cross-section (sc). Expression in the neural tube is maintained at E17.5 and also seen in the olfactory bulbs (ob) in some embryos. (E) Transgenic reporter embryos carrying agCNE6 only show restricted expression in part of the hindbrain (hb), particularly at the midbrain-hindbrain boundary (mhb) and along the length of the spinal cord (sc). |
| Gene: | |
|---|---|
| Fish: | |
| Anatomical Terms: | |
| Stage Range: | Prim-5 to Protruding-mouth |
Reprinted from Developmental Biology, 387(2), Bhatia, S., Monahan, J., Ravi, V., Gautier, P., Murdoch, E., Brenner, S., van Heyningen, V., Venkatesh, B., and Kleinjan, D.A., A survey of ancient conserved non-coding elements in the PAX6 locus reveals a landscape of interdigitated cis-regulatory archipelagos, 214-228, Copyright (2014) with permission from Elsevier. Full text @ Dev. Biol.