Fig. 1
|
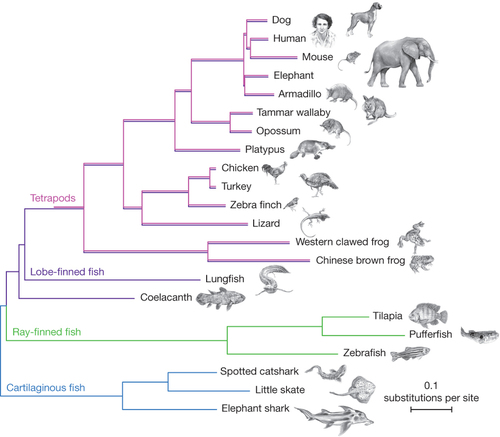
Multiple sequence alignments of 251 genes with a 1:1 ratio of orthologues in 22 vertebrates and with a full sequence coverage for both lungfish and coelacanth were used to generate a concatenated matrix of 100,583 unambiguously aligned amino acid positions. The Bayesian tree was inferred using PhyloBayes under the CAT +GTR+ Γ4 model with confidence estimates derived from 100 gene jack-knife replicates (support is 100% for all clades but armadillo + elephant with 45%). The tree was rooted on cartilaginous fish, and shows that the lungfish is more closely related to tetrapods than the coelacanth, and that the protein sequence of coelacanth is evolving slowly. Pink lines (tetrapods) are slightly offset from purple lines (lobe-finned fish), to indicate that these species are both tetrapods and lobe-finned fish. |