Fig. S11
|
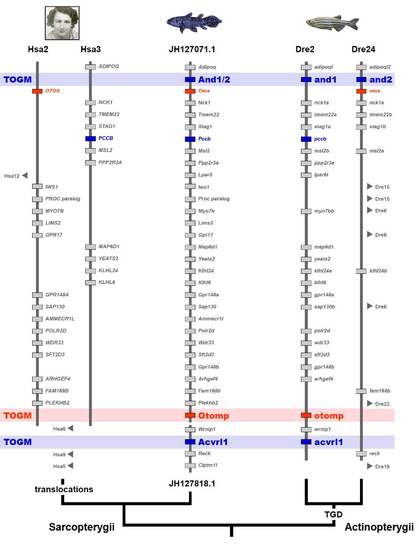
Evolution of the And1/2 – Otomp – Acvrl1 region in bony vertebrates. Orthologs of genes on coelacanth scaffolds JH126651.1 and JH127818.1 are distributed across chromosomes Hsa2 and Hsa3 in the human (e.g. M. Courtenay-Latimer) genome, indicating translocations on the tetrapod branch leading to human, while teleost (co-) orthologs of these Latimeria genes are distributed among two zebrafish chromsomes, Dre2 and Dre24, which contain paralogons from the teleost genome duplication (TGD). Note that gene order in human and zebrafish is presented according to the coelacanth gene order. The region contains several genes involved in fin (blue) and ear development (red), among them three genes lost in tetrapods: Zebrafish actinodin 1 (and1) and actinodin2 (and2) genes encode structural proteins of the actinotrichia, the skeletal elements that stiffen fin folds, and their loss in tetrapods has been suggested to have contributed to the fin-to-limb transition180. Loss of acvrl1, encoding a BMP receptor, leads in teleosts to the malformation of the ventral tail fin (lost-a-fin mutant)211-212. The otolith matrix protein (otomp) gene is essential for otolith formation in the zebrafish ear213, the tetrapod homolog of which evolved adaptations for signal detection in air. |