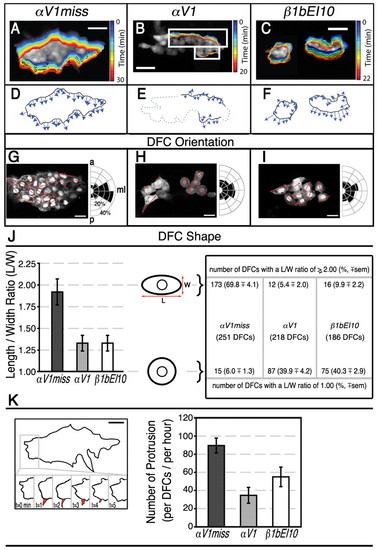
Effects of integrin αV1 or β1b knockdown on DFC orientation, shape and protrusive activity. At mid-gastrulation stages, DFCs show dynamic cellular protrusive activity and only control DFCs are normally mediolaterally oriented and have elongated morphology. (A-C) Dorsal views of Tg(sox17:GFP)-expressing embryos. Time-lapse images of migratory DFCs were collected by confocal microscopy at mid-gastrulation and migration was highlighted with pseudo-colors at one-minute intervals at the DFC cluster edge. Pseudo-colored outlines show the edges of DFC clusters at the timepoints indicated on the right. (A) At 70% E, all DFCs were clustered in αV1miss control embryos (1.75 ng; n=10) and remained clustered until the end of gastrulation (see also Movie 1 in the supplementary material). (B) Some DFCs in αV1 morphants (1.25 ng; n=6) formed clusters (white rectangles). Later in development, cells outside of these clusters detached from each other (see also Movie 3 in the supplementary material). (C) β1bEI10 (5 ng; n=6) morphants had multiple DFC clusters (see also Movie 5 in the supplementary material). (D-F) Pseudopod-like protrusion behavior of representative DFC cluster edges is shown between 4 to 5 minutes. Vectors illustrate the relative protrusion velocities of cells at the cluster edge and the direction of protrusions. Each vector represents a protrusion event, where its speed is proportional to the vector length. (D) Protrusions in αV1miss morphant DFCs formed towards the vegetal pole, whereas DFC protrusions in αV1 (E) and β1bEI10 morphants (F) lost their directionality. The relative locations of αV1 morphant DFCs that did not form any clusters, or detached from each other later in development, are highlighted with dotted lines (E). (G-I) Representative ∼3 μm thick focal plane confocal images of Tg(sox17:GFP)-expressing DFC clusters. Dorsal views of 80% E embryos are shown in all panels, anterior to the top. Half-rose diagrams show the angular distribution of the long axis of individual DFCs with respect to anterior-posterior (a,p) axis. Red arrows indicate the orientation of the long axis in each individual DFC that showed clear cellular boundary at these representative focal planes. Dividing DFCs (asterisks) were excluded from these measurements. ml, mediolateral axis. (J) The length (L) and width (W) measurements of individual DFCs were used to establish L-W ratios ± s.e.m. The majority of control αV1miss-injected embryos had a L-W ratio of 2.00, represented with ellipsoid cell shape. However, the L-W ratio in a significant fraction of morphant DFCs was close to 1.00, represented with circular cell shape. Total number of embryos and DFCs scored to determine individual cell orientation, length and width (L/W) ratios: 1.5 ng αV1miss (n=11, 251 DFCs); 1.25 ng αV1 (n=11, 218 DFCs); 5 ng β1bEI10 (n=11, 186 DFCs). (K) Average number of new and discrete large protrusions developed per DFC per hour were manually counted from the time-lapse confocal microscopy of migratory DFCs. DFC cluster edge in an αV1miss-injected embryo (see Movie 1 in the supplementary material) is outlined and a representative large protrusion is highlighted in red. The behavior and life span of the protrusions are shown below from 0-5 minutes. The bar graph shows the effects of αV and β1b knockdown on the number of protrusions formed per DFC per embryo per hour ± s.e.m. Total number of embryos studied: 1.5 ng αV1miss (n=5); 1.25 ng αV1 (n=3); 5 ng β1bEI10 (n=3). Scale bars: 50 μm in A-F; 20 μm G-I,K.
|

