Fig. S1
- ID
- ZDB-FIG-090817-20
- Publication
- McMahon et al., 2009 - Lmx1b is essential for survival of periocular mesenchymal cells and influences Fgf-mediated retinal patterning in zebrafish
- Other Figures
- All Figure Page
- Back to All Figure Page
|
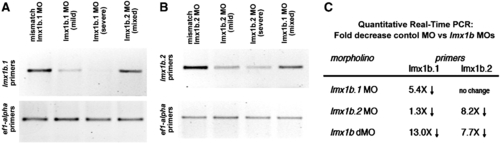
Analysis of transcript levels of lmx1b.1 and lmx1b.2 following morpholino knock-down. Ethidium bromide stained gel of RT-PCR analysis of either (A) lmx1b.1 or (B) lmx1b.2. For each, the cDNA source is shown at the top and amplification from ef1-alpha primers is shown as an input control. With either lmx1b.1 or lmx1b.2 morpholinos, we did not detect alternate splice bands, but did find depletion of the normally-spliced transcript. For lmx1b primers 35 cycles were carried out, while reactions with ef1-alpha were amplified for 20 cycles. Mild and Severe refers to the degree of the phenotype as defined in the Materials and methods. Mixed indicates equal numbers of mild and severe. (C) Fold change in transcript levels from 18 somite stage embryos as assessed by quantitative real-time RT-PCR. |
Reprinted from Developmental Biology, 332(2), McMahon, C., Gestri, G., Wilson, S.W., and Link, B.A., Lmx1b is essential for survival of periocular mesenchymal cells and influences Fgf-mediated retinal patterning in zebrafish, 287-298, Copyright (2009) with permission from Elsevier. Full text @ Dev. Biol.