- Title
-
Epithelial transcription factor Elf3 mediates host immune responses to microbiota and protects against aerocystitis in zebrafish
- Authors
- Davis, B.R., Lickwar, C.R., Löhr, C.V., Wen, J., Morash, M., Sweeney, M.I., Reich, E.L., Moore, P.J., Tobin, D.M., Rawls, J.F.
- Source
- Full text @ MBio
|
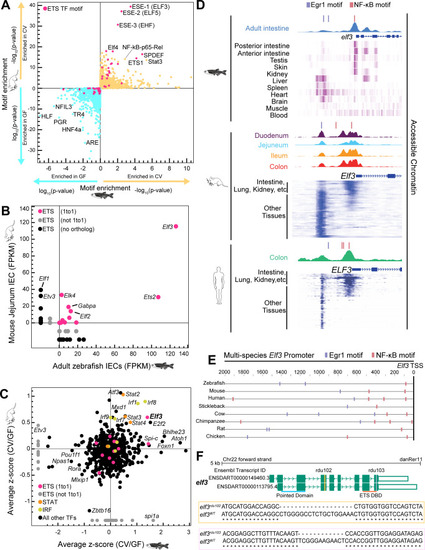
Identification of a candidate TF mediating conserved host responses to the microbiota. ( |
|
|
|
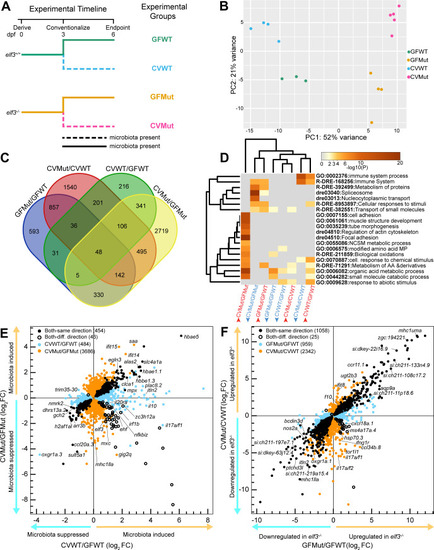
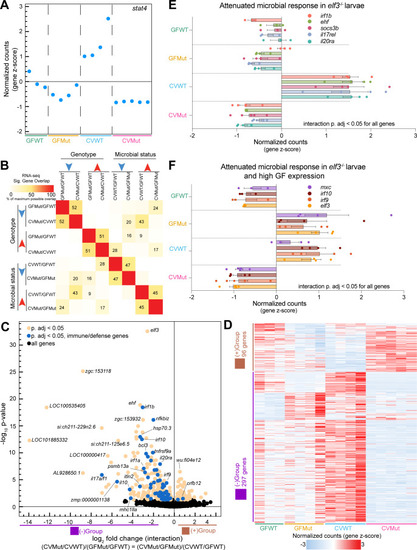
Identification of interaction genes that integrate |
|
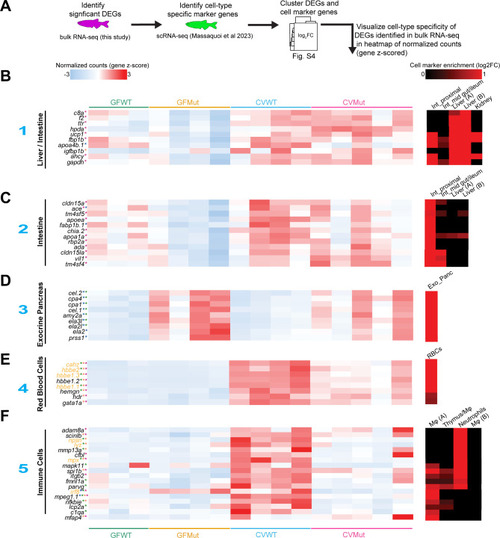
Differentially expressed genes in larval RNA-seq exhibit cell-type specificity. ( |
|
|
|
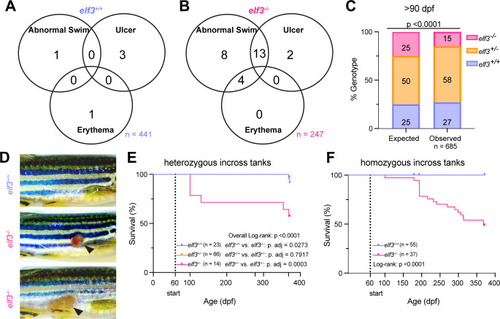
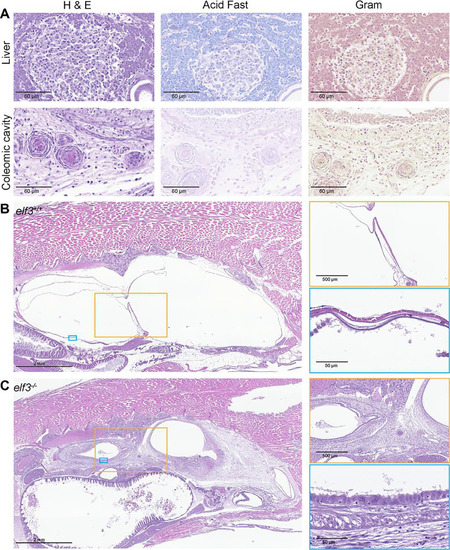
Moribund |
|
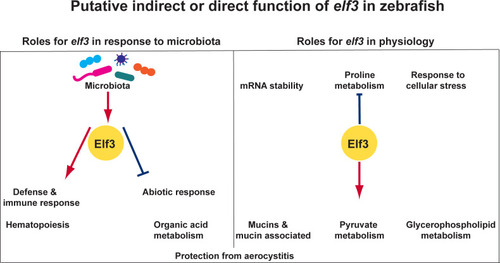
Summary model indicating putative roles for elf3 in zebrafish. |