- Title
-
Oxidative phosphorylation is required for cardiomyocyte re-differentiation and long-term fish heart regeneration
- Authors
- Lekkos, K., Hu, Z., Nguyen, P.D., Honkoop, H., Sengul, E., Alonaizan, R., Koth, J., Ying, J., Lemieux, M.E., Kenward, A., Keeley, S., Spanjaard, B., Kennedy, B.W.C., Sun, X., Banecki, K., Potts, H.G., Ruggiero, G., Montgomery, J., Panáková, D., Junker, J.P., Heather, L.C., Wang, X., Gonzalez-Rosa, J.M., Bakkers, J., Mommersteeg, M.T.M.
- Source
- Full text @ Nat Cardiovasc Res
|
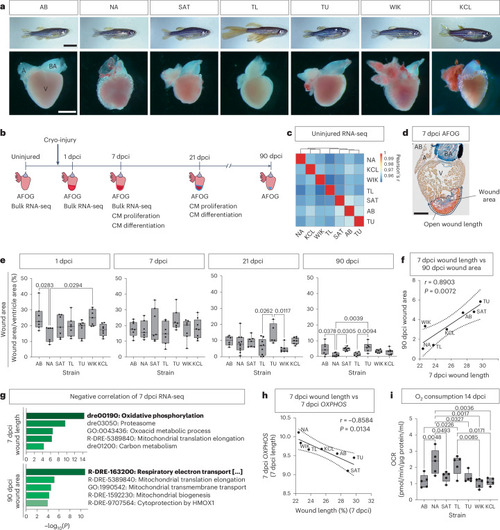
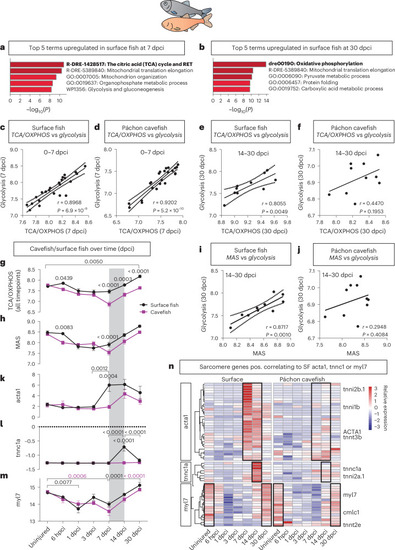
Differential regenerative response to cryo-injury among wild-type adult zebrafish strains identifies OXPHOS as beneficial for regeneration. |
|
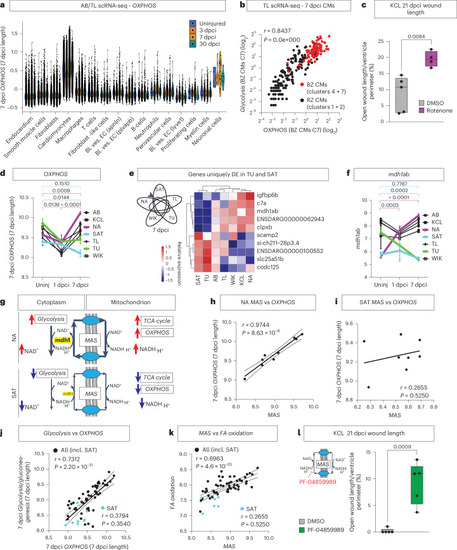
Upregulation of the MAS drives |
|
OXPHOS is not required for cardiomyocyte proliferation. |
|
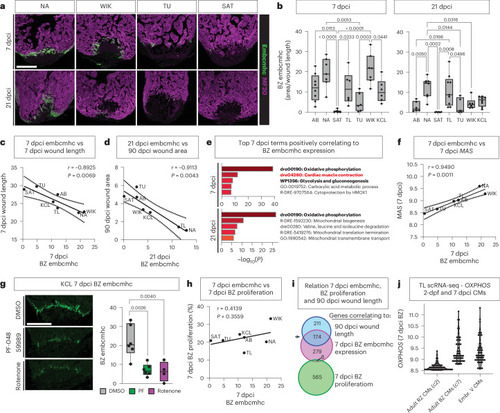
Levels of |
|
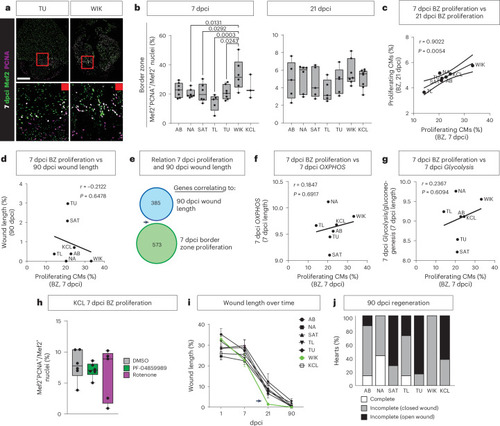
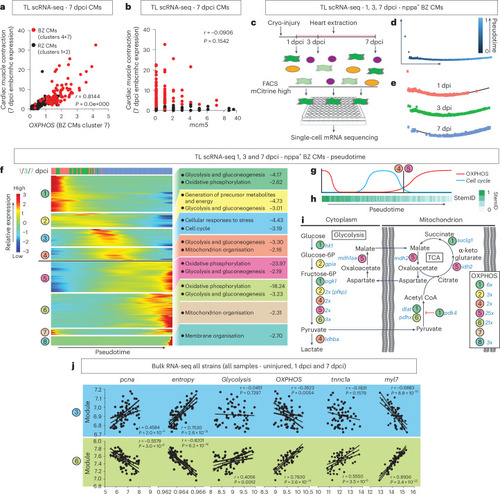
Cardiomyocyte proliferation is separated in time from OXPHOS and cardiomyocyte re-differentiation. |
|
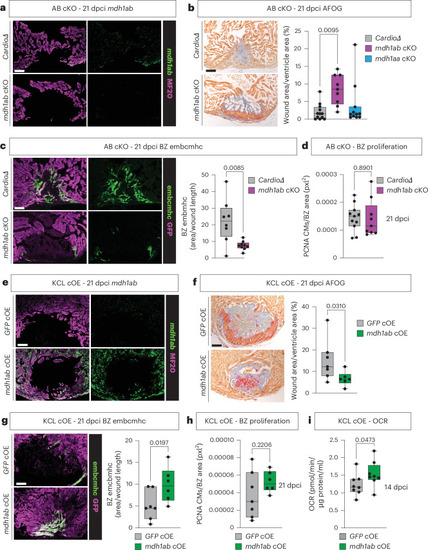
Genetic manipulation of |
|
Strongly reduced re-differentiation gene expression in |