- Title
-
Direct lysine dimethylation of IRF3 by the methyltransferase SMYD3 attenuates antiviral innate immunity
- Authors
- Wang, Z., Chen, X., Zhu, C., Fan, S., Tang, J., Deng, H., Sun, X., Liu, X., Xiao, W.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
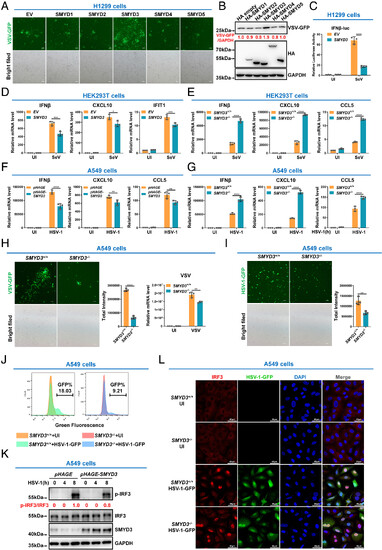
SMYD3 attenuates the cellular antiviral immune response. (A and B) The effect of ectopic expression of SMYDs on VSV-GFP replication in H1299 cells. (C) Luciferase activity of IFNβ promoter reporter in H1299 cells transfected with empty pCMV vector (EV) or pCMV-SMYD3 for 24 h, followed by uninfected (UI) or infected with SeV for 8 h. (D) qPCR analysis of IFNβ, CXCL10, and IFIT1 mRNA in HEK293T cells transfected with empty pCMV EV or pCMV-SMYD3, followed by UI or infected with SeV. (E) qPCR analysis of IFNβ, CXCL10, and CCL5 mRNA in SMYD3-intact (SMYD3+/+) or SMYD3-null (SMYD3−/−) HEK293T cells. (F) qPCR analysis of IFNβ, CXCL10, and CCL5 mRNA in A549 cells infected with pHAGE lentivirus (control) or pHAGE-SMYD3 lentivirus, followed by UI or infected with HSV-1 for 8 h. (G) qPCR analysis of IFNβ, CXCL10, and CCL5 mRNA in SMYD3-intact (SMYD3+/+) or SMYD3-null (SMYD3−/−) A549 cell UI or infected with HSV-1 for 8 h. (H) Microscopic imaging (Left panels) and viral mRNA detection (Right panels) of VSV replication in SMYD3-intact (SMYD3+/+) or SMYD3-null (SMYD3−/−) A549 cells. (I) Microscopy imaging of HSV-1-GFP virus replication in SMYD3-intact (SMYD3+/+) or SMYD3-null (SMYD3−/−) A549 cells. (J) Flow cytometric analysis of HSV-1 GFP virus replication in SMYD3-intact (SMYD3+/+) or SMYD3-null (SMYD3−/−) A549 cells UI or infected with HSV-1-GFP virus for 8 h. (K) Phosphorylation of IRF3 in A549 cells infected with pHAGE lentivirus (control) or pHAGE-SMYD3 lentivirus, followed by HSV-1 infection for 0, 4, and 8 h. Quantification for phosphorylation of IRF3 was shown in red. (L) Microscopic imaging of IRF3 localization in SMYD3-intact (SMYD3+/+) or SMYD3-null (SMYD3−/−) A549 cells UI or infected with HSV-1-GFP for 8 h. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Graphs represent fold induction relative to the untreated cells. Data are presented as the mean values of a representative experiment performed in triplicate (C–G, and H) or as representative data (A, B, H, I, J, and L); these experiments were repeated independently at least three times, and error bars indicate S.D. |
|
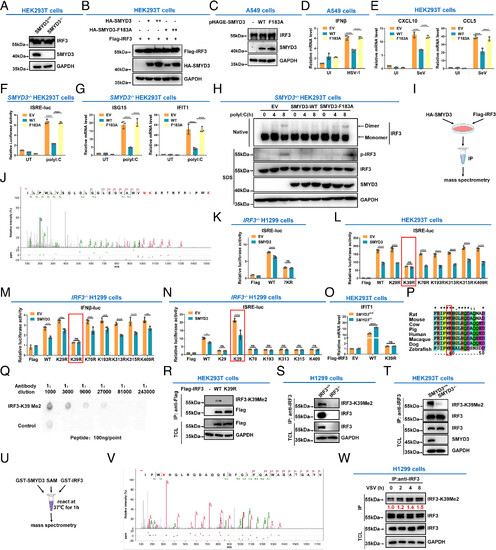
SMYD3 catalyzes the methylation of IRF3 at lysine 39. (A) Immunoblotting of endogenous IRF3 in SMYD3+/+ or SMYD3−/− HEK293T cells. (B) Immunoblotting of ectopic IRF3 protein in HEK293T cells transfected with WT SMYD3 or the enzymatically inactive mutant of SMYD3 (F183A) at increasing doses. (C) Immunoblotting of endogenous IRF3 in A549 cells infected with pHAGE-SMYD3, or pHAGE-SMYD3(F183A) lentiviruses. (D) qPCR analysis of IFNβ in A549 cells transfected with empty vector (EV), WT SMYD3 (WT), or the enzymatically inactive mutant of SMYD3 (F183A), followed by UI or infected with HSV-1 for 8 h. (E) qPCR analysis of CXCL10 and CCL5 in HEK293T cells transfected with empty vector (EV), WT SMYD3 (WT), or the enzymatically inactive mutant of SMYD3 (F183A), followed by UI or infected with SeV for 8 h. (F) Luciferase activity of the ISRE reporter in SMYD3−/− HEK293T cells transfected with empty vector (EV), WT SMYD3 (WT), or the enzymatically inactive mutant of SMYD3 (F183A), followed by untransfected (UT) or transfected with poly I:C for 8 h. (G) qPCR analysis of ISG15 and IFIT1 in SMYD3−/− HEK293T cells transfected with empty vector (EV), WT SMYD3 (WT), or the enzymatically inactive mutant of SMYD3 (F183A), followed by untransfected (UT) or transfected with poly I:C for 8 h. (H) Phosphorylation and dimerization of IRF3 in SMYD3−/− HEK293T cells transfected with empty vector (EV), WT SMYD3 (WT), or the enzymatically inactive mutant of SMYD3 (F183A), followed by untransfected (UT) or transfected with poly I:C for 0, 4, and 8 h. (I) Scheme of detection of IRF3 methylation by mass spectrometry analysis. (J) Mass spectrometry analysis revealed that lysine 39 in IRF3 was dimethylated by SMYD3. (K) Luciferase activity of the ISRE reporter in IRF3−/− H1299 cells transfected with SMYD3 together with WT IRF3 (WT) or the lysine to arginine mutant (all the seven lysine residues, including K29, K39, K70, K193, K313, K315, and K409, were simultaneously mutated to arginine residues). (L) Luciferase activity of the ISRE reporter in HEK293T cells transfected with SMYD3 together with WT IRF3 (WT) or the lysine to arginine mutants, including K29R, K39R, K70R, K193R, K313R, K315R, or K409R. (M) Luciferase activity of the IFNβ promoter reporter in IRF3−/− H1299 cells transfected SMYD3 together with WT IRF3 (WT), or its lysine to arginine mutants. (N) Luciferase activity of the ISRE reporter in IRF3−/− H1299 cells transfected with SMYD3 together with WT IRF3 (WT), or its lysine to arginine mutants, including K29, K39, K70, K193, K313, K315, or K409 (In which only K29, K39, K70, K193, K313, K315, or K409, was kept intact respectively and other six lysine residues were simultaneously mutated to arginine residues). (O) qPCR analysis of IFIT1 in SMYD3+/+ and SMYD3−/− HEK293T cells transfected with empty vector (EV), WT IRF3 (WT), or IRF3-K39R mutant. (P) Sequence alignment of partial IRF3 (34-50 amino acids) from human, mouse, rat, cow, pig, macaque, dog, and zebrafish. The red box indicates a conserved lysine (K39). (Q) Dot blot assay for the specificity of the anti-IRF3-K39Me2 antibody. Equal amounts of dimethylated peptides or the control peptides were immunoblotted with the indicated dilutions of anti-IRF3-K39Me2 antibody. (R) HEK293T cells were transfected with empty Flag vector (−), Flag-IRF3 (WT), or Flag-IRF3-K39R, followed by immunoprecipitating with anti-Flag antibody, and immunoblotting with anti-IRF3-K39Me2 antibody. (S) Cell lysates from IRF3-intact or IRF3-null H1299 cells (IRF3+/+ or IRF3−/−) were extracted, followed by immunoprecipitation with anti-IRF3 antibody, and immunoblotting with anti-IRF3 and anti-IRF3-K39Me2 antibodies. (T) Cell lysates from SMYD3-intact or SMYD3-null HEK293T cells (SMYD3+/+ or SMYD3−/−) were extracted, followed by immunoprecipitation with anti-IRF3 antibody, and immunoblotting with anti-IRF3 and anti-IRF3-K39Me2 antibodies. (U) Scheme for the in vitro methylation assay and detection of IRF3 methylation by mass spectrometry analysis. (V) Mass spectrometry analysis revealed that lysine 39 in IRF3 was methylated by bacterially expressed SMYD3. (W) IRF3 methylation at lysine 39 was steadily increased after VSV infection. Anti-IRF3 antibody was used for Co-IP, and anti-IRF3-K39Me2 antibody was used for detection. IP, immunoprecipitation; TCL, total cell lysate; empty vector (EV); uninfected (UI); UT, untransfected; ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Graphs represent fold induction relative to the untreated cells. Data are presented as the mean values of a representative experiment performed in triplicate (D–G and K–O) or as representative data (A–C, H, R–T, and W); these experiments were repeated independently at least three times, and error bars indicate S.D. |
|
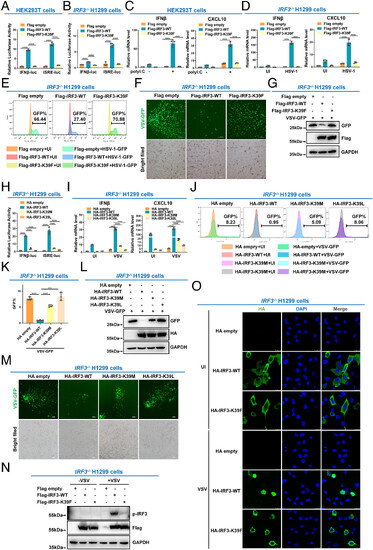
IRF3 is inactivated by methylation at lysine 39. (A) Luciferase activity of IFNβ promoter reporter and ISRE reporter in HEK293T cells transfected with empty Flag vector, WT IRF3 (Flag-IRF3-WT), or the methylation mimic mutant (Flag-IRF3-K39F). (B) Luciferase activity of the IFN-β promoter reporter and ISRE reporter in IRF3−/− H1299 cells transfected with empty Flag vector, WT IRF3 (Flag-IRF3-WT), or the methylation mimic mutant (Flag-IRF3-K39F). (C) qPCR analysis of IFNβ and CXCL10 mRNA in HEK293T cells transfected with empty Flag vector, WT IRF3 (Flag-IRF3-WT), or the methylation mimic mutant (Flag-IRF3-K39F), followed by transfection with (+) or without (−) poly I: C. (D) qPCR analysis of IFNβ and CXCL10 mRNA in IRF3−/− H1299 cells transfected with empty Flag vector, WT IRF3 (Flag-IRF3-WT), or the methylation mimic mutant (Flag-IRF3-K39F), followed by UI or infected with HSV-1. (E) Flow cytometric analysis of HSV-1-GFP virus replication in IRF3−/− H1299 cells transfected with empty Flag vector, WT IRF3 (Flag-IRF3-WT), or the methylation mimic mutant (Flag-IRF3-K39F), followed by UI or infected with HSV-1-GFP virus. (F) Microscopic imaging of VSV-GFP virus replication in IRF3−/− H1299 cells transfected with empty Flag vector, WT IRF3 (Flag-IRF3-WT), or the methylation mimic mutant (Flag-IRF3-K39F), followed by infection with VSV-GFP virus. (G) Immunoblotting analysis of VSV-GFP virus replication in IRF3−/− H1299 cells transfected with empty Flag vector, WT IRF3 (Flag-IRF3-WT), or the methylation mimic mutant (Flag-IRF3-K39F), followed by infection with VSV-GFP virus. (H) Luciferase activity of the IFN-β promoter reporter and ISRE reporter in IRF3−/− H1299 cells transfected with empty HA vector, WT IRF3 (HA-IRF3-WT), or the methylation mimic mutant (HA-IRF3-K39M or HA-IRF3-K39L mutant). (I) qPCR analysis of IFNβ and CXCL10 mRNA in IRF3−/− H1299 cells transfected with empty HA vector, WT IRF3 (HA-IRF3-WT), or the methylation mimic mutant (HA-IRF3-K39M or HA-IRF3-K39L mutant), followed by UI or infected with VSV. (J and K) Flow cytometric analysis of VSV-GFP virus replication in IRF3−/− H1299 cells transfected with empty HA vector, WT IRF3 (HA-IRF3-WT), or the methylation mimic mutant (HA-IRF3-K39M or HA-IRF3-K39L mutant), followed by UI or infected with VSV-GFP virus. (L) Immunoblotting analysis of VSV-GFP virus replication in IRF3−/− H1299 cells transfected with empty HA vector, WT IRF3 (HA-IRF3-WT), or the methylation mimic mutant (HA-IRF3-K39M or HA-IRF3-K39L mutant), followed by infection with VSV-GFP virus. (M) Microscopic imaging of VSV-GFP virus replication in IRF3−/− H1299 cells transfected with empty HA vector, WT IRF3 (HA-IRF3-WT), or the methylation mimic mutant (HA-IRF3-K39M or HA-IRF3-K39L mutant), followed by infection with VSV-GFP virus. (N) Phosphorylation of IRF3 in IRF3−/− H1299 cells transfected with empty Flag vector, WT IRF3 (Flag-IRF3-WT), or the methylation mimic mutant (Flag-IRF3-K39F), followed by VSV infection for 8 h.(O) Localization of WT IRF3 (HA-IRF3-WT) and the methylation mimic mutant (HA-IRF3-K39F) in IRF3−/− H1299 cells UI or infected with VSV. ****P < 0.0001. Graphs represent fold induction relative to the untreated cells. Data are presented as the mean values of a representative experiment performed in triplicate (A–D, H, I, and K) or as representative data (E–G, J, and L–O); these experiments were repeated independently at least three times, and error bars indicate S.D. |
|
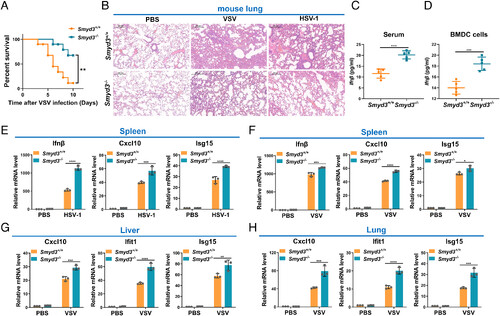
Disruption of Smyd3 in mice results in increased susceptibility to lethal viral infection. (A) Survival (Kaplan–Meier curve) of Smyd3+/+ (n = 10) and Smyd3−/− (n = 10) mice by intraperitoneal injection of VSV (1 × 107 plaque-forming units (PFU)/ per mouse) and monitored for 10 d. **P < 0.01. (B) Hematoxylin and eosin-stained images of lung sections from Smyd3+/+ or Smyd3−/− mice injected intraperitoneally with phosphate buffer saline (PBS) (vehicle control), VSV (1 × 107 PFU/per mouse) or HSV-1 (5 × 107 PFU/per mouse) for 24 h. (C) ELISA of Ifnβ in the serum of Smyd3+/+ or Smyd3−/− mice injected intraperitoneally with VSV (1 × 107 PFU/per mouse) for 24 h. (D) ELISA of Ifn-β in the supernatant of Smyd3+/+ or Smyd3−/− BMDCs infected with VSV for 8 h. (E) qPCR analysis of Ifnβ, Cxcl10, and Isg15 mRNA in spleens of Smyd3+/+ or Smyd3−/− mice injected intraperitoneally with PBS or HSV-1 (5 × 107 PFU/per mouse) for 24 h. (F) qPCR analysis of Ifnβ, Cxcl10, and Isg15 mRNA in spleens of Smyd3+/+ or Smyd3−/− mice injected intraperitoneally with PBS or VSV (1 × 107 PFU/per mouse) for 24 h. (G) qPCR analysis of Cxc10, Ifit1, and Isg15 mRNA in the liver of Smyd3+/+ or Smyd3−/− mice injected intraperitoneally with PBS or VSV (1 × 107 PFU/per mouse) for 24 h. (H) qPCR analysis of Cxcl10, Ifit1, and Isg15 mRNA in the lungs of Smyd3+/+ or Smyd3−/− mice injected intraperitoneally with PBS or VSV (1 × 107 PFU/per mouse) for 24 h. PBS, phosphate buffer saline; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Data are presented as the mean values of a representative experiment performed in triplicate (E–H) or as representative data (B); these experiments were repeated independently at least three times, and error bars indicate S.D. |
|
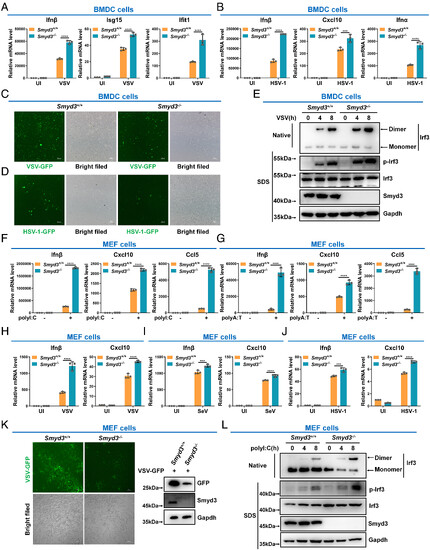
Loss of Smyd3 enhances the cellular antiviral immune response. (A) qPCR analysis of Ifnβ, Isg15, and Ifit1 mRNA in Smyd3+/+ and Smyd3−/− BMDCs UI or infected with VSV for 8 h. (B) qPCR analysis of Ifnβ, Cxcl10, and Ifnα mRNA in Smyd3+/+ and Smyd3−/− BMDCs UI or infected with HSV-1 for 8 h. (C) Microscopic imaging of VSV-GFP virus replication in Smyd3+/+ and Smyd3−/− BMDCs. (D) Microscopic imaging of HSV-1-GFP virus replication in Smyd3+/+ and Smyd3−/− BMDCs. (E) Phosphorylation and dimerization of Irf3 in Smyd3+/+ and Smyd3−/− BMDCs infected with VSV for 0, 4, and 8 h. (F) qPCR analysis of Ifnβ, Cxcl10, and Ccl5 mRNA in Smyd3+/+ and Smyd3−/− MEF cells transfected with (+) or without (−) poly I:C for 8 h. (G) qPCR analysis of Ifnβ, Cxcl10, and Ccl5 mRNA in Smyd3+/+ and Smyd3−/− MEF cells transfected with (+) or without (−) poly A:T for 8 h. (H) qPCR analysis of Ifnβ and Cxcl10 mRNA in Smyd3+/+ and Smyd3−/− MEF cells UI or infected with VSV for 8 h. (I) qPCR analysis of Ifnβ and Cxcl10 mRNA in Smyd3+/+ and Smyd3−/− MEF cells UI or infected with SeV for 8 h. (J) qPCR analysis of Ifnβ and Cxcl10 mRNA in Smyd3+/+ and Smyd3−/− MEF cells UI or infected with HSV-1 for 8 h. (K) Microscopic imaging (Left panels) and immunoblotting (Right panels) of VSV-GFP virus replication in Smyd3+/+ and Smyd3−/− MEF cells. (L) Phosphorylation and dimerization of Irf3 in Smyd3+/+ and Smyd3−/− MEF cells transfected with poly I:C for 0, 4, and 8 h. ***P < 0.001 and ****P < 0.0001. Graphs represent fold induction relative to the untreated cells. Data are presented as the mean values of a representative experiment performed in triplicate (A, B, and F–J) or as representative data (C–E, K, and L); these experiments were repeated independently at least three times, and error bars indicate S.D. |