- Title
-
Induced formation of primordial germ cells from zebrafish blastomeres by germplasm factors
- Authors
- Wang, X., Zhu, J., Wang, H., Deng, W., Jiao, S., Wang, Y., He, M., Zhang, F., Liu, T., Hao, Y., Ye, D., Sun, Y.
- Source
- Full text @ Nat. Commun.
|
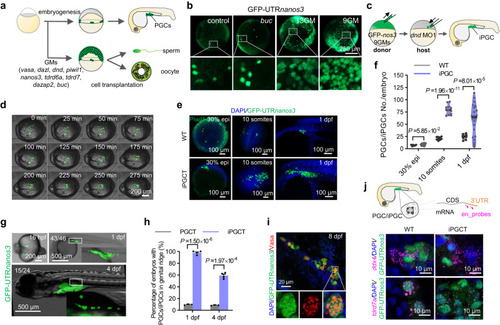
Generation of iPGC via germplasm in vivo. |
|
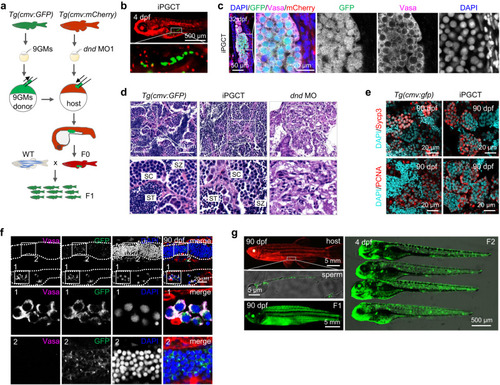
Functional gametes were obtained using iPGCT. |
|
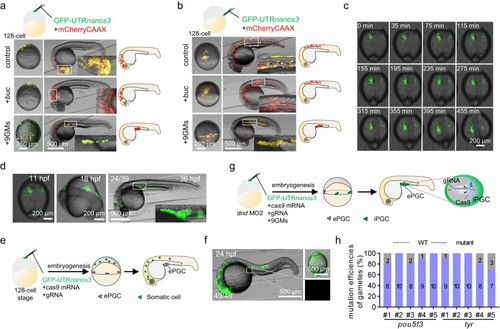
Functional gametes were produced by the single blastomere induction of iPGCs. Injection of 9GM or |
|
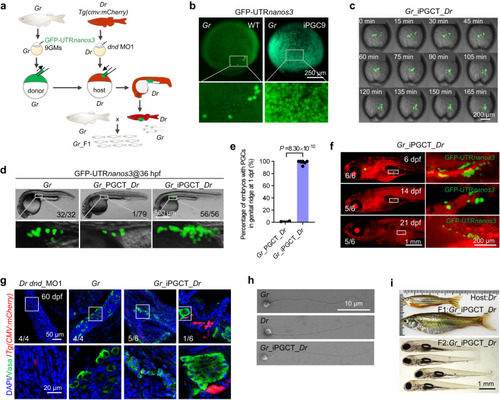
Generation of xenogametes by iPGCT. |
|
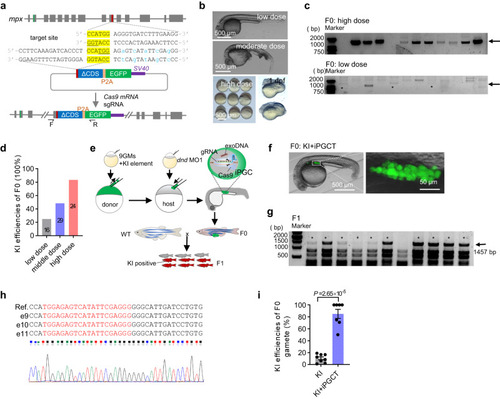
iPGCT greatly improved the knock-in (KI) efficiency of gametes. |