- Title
-
An injury-responsive mmp14b enhancer is required for heart regeneration
- Authors
- Zlatanova, I., Sun, F., Wu, R.S., Chen, X., Lau, B.H., Colombier, P., Sinha, T., Celona, B., Xu, S.M., Materna, S.C., Huang, G.N., Black, B.L.
- Source
- Full text @ Sci Adv
|
( |
|
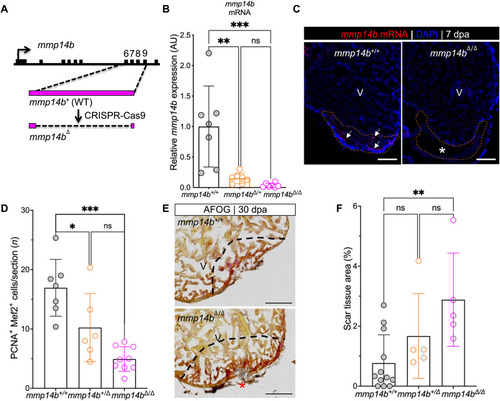
Inactivation of the ( |
|
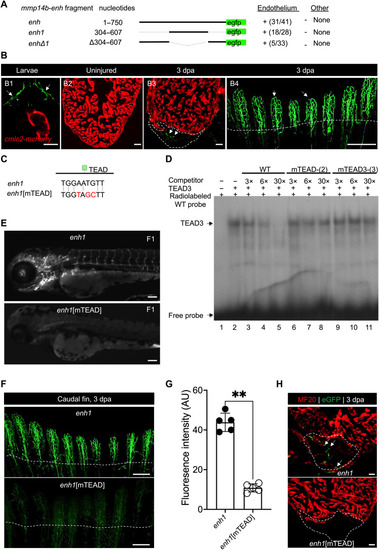
Identification of an injury-responsive ( |
|
Identification of a TEAD motif–dependent core region of ( |
|
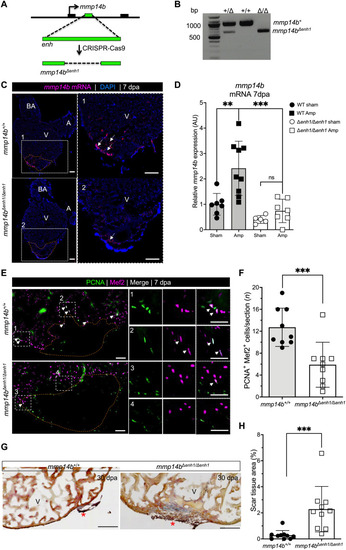
( |
|
( |
|
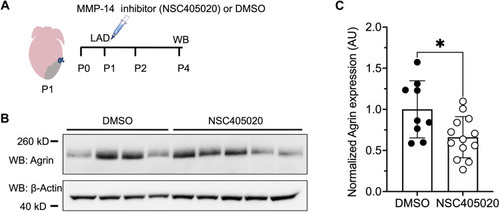
MMP-14 activity facilitates Agrin availability in the extracellular matrix of neonatal mice. ( |