- Title
-
Ergolide mediates anti-cancer effects on metastatic uveal melanoma cells and modulates their cellular and extracellular vesicle proteomes
- Authors
- Sundaramurthi, H., Tonelotto, V., Wynne, K., O'Connell, F., O'Reilly, E., Costa-Garcia, M., Kovácsházi, C., Kittel, A., Marcone, S., Blanco, A., Pallinger, E., Hambalkó, S., Piulats Rodriguez, J.M., Ferdinandy, P., O'Sullivan, J., Matallanas, D., Jensen, L.D., Giricz, Z., Kennedy, B.N.
- Source
- Full text @ Open Res Eur
|
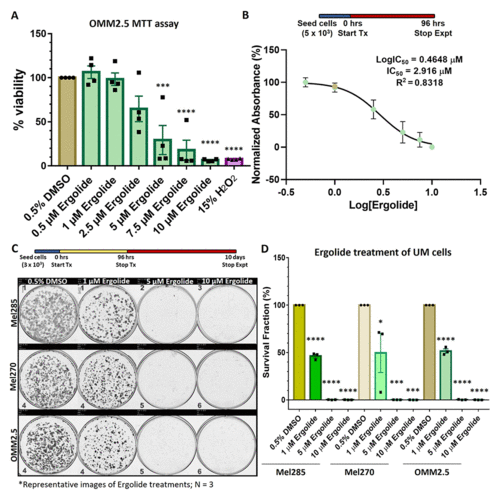
Ergolide treatment significantly reduced UM/MUM cell viability. (A, B) Dose-response analysis and determination of IC50 of ergolide in OMM2.5 cells as 2.9 μM via the MTT assay. (C, D) Long-term survival of Mel285, Mel270 and OMM2.5 clones were significantly (****, adjusted p=0.0001) reduced in ergolide treated samples compared to vehicle control (N = 3). Statistical analysis performed using One-way ANOVA with Dunnett's Test for Multiple Comparisons. |
|
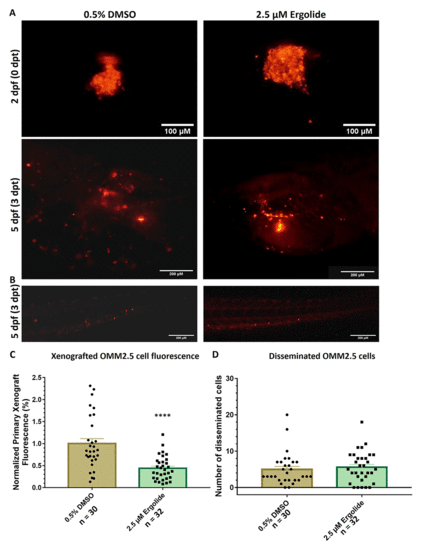
Anti-cancer effects of ergolide observed in vivo in zebrafish OMM2.5 xenograft models. (A, C) Representative images of zebrafish larvae transplanted with OMM2.5Dil labelled cells at 2 days post fertilization (dpf, top panel); representative images of zebrafish larvae transplanted with OMM2.5Dil labelled cells at 5 dpf, 3 days post treatment (dpt, bottom panel). A 56% (****, p<0.0001%) reduction, on average, in primary xenograft fluorescence observed in the 2.5 μM ergolide (n = 32) treated samples compared to 0.5% DMSO (n = 30) treated samples. (B, D) A significant difference was not detected in the average number of disseminated cells. Statistical analysis performed using Student's T-test. |
|
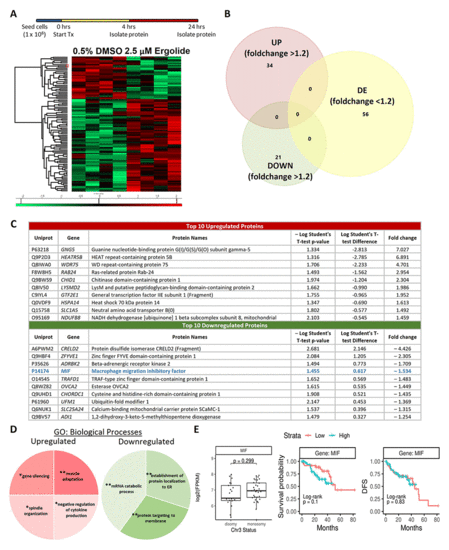
Proteome profiling following 24 hours ergolide treatment of OMM2.5 cells. (A) Schematic diagram depicting treatment regime and heatmap highlighting differentially expressed proteins. (B) Venn diagram showing the number of proteins significantly differentially expressed (up/down) or differentially expressed (DE), given a cutoff of p≤0.05 and a ≥ 1.2 fold change. (C) Table of top ten proteins that are significantly up- or down-regulated following 24 hours of ergolide treatment. (D) Pathways enriched for GO: Biological processes. (E) TCGA analysis of UM patient samples comparing MIF expression levels to chromosome 3 (Chr 3) status, overall survival and disease-free survival (DFS), (n = 80). |
|
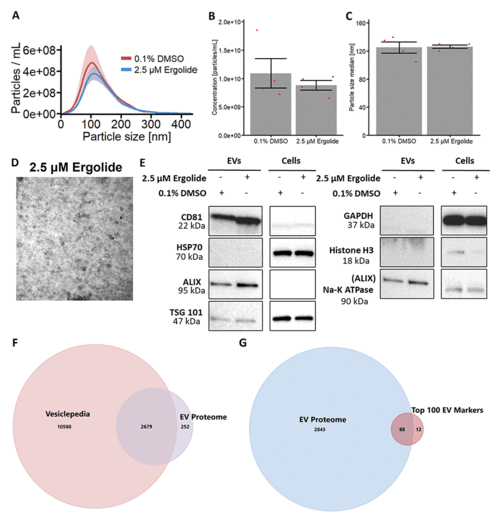
Characterization of EV isolated from OMM2.5 cells. (A) NTA analysis of isolated OMM2.5 EV particle size ranging from 50 nm to 250 nm in both 0.1% treated DMSO and 2.5 μM ergolide treated samples. (B and C) A significant difference in the amount of particles and particle size was not observed between ergolide treated samples and vehicle control. (D) TEM image of particles isolated from OMM2.5 cells. (E) Immunoblot analysis of select proteins in OMM2.5 EV isolates and cell lysate (N = 1). (F) Comparison between Vesiclepedia compendium and EV proteome. (G) 88 proteins from the list of top 100 EV markers identified in OMM2.5 EV proteome. |
|
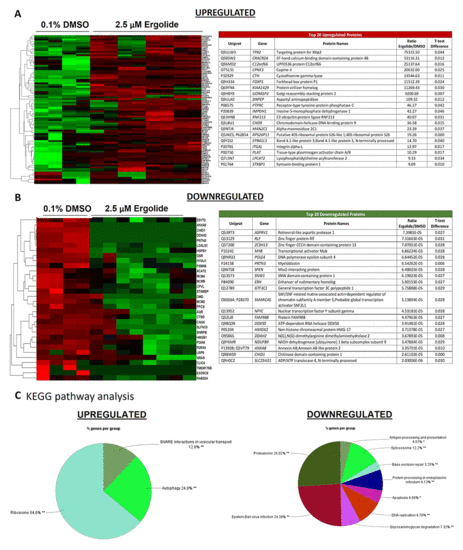
Analysis of differentially expressed proteins in EVs isolated from OMM2.5 cells treated with ergolide. (A) Heatmap of upregulated proteins in ergolide treatment group (left panel); list of top 20 significantly upregulated proteins in samples treated with ergolide (right panel). (B) Heatmap of downregulated proteins in ergolide treatment group (left panel); list of top 20 significantly downregulated proteins in samples treated with ergolide (right panel). (C) KEGG pathway analysis to determine pathways enriched in EVs following ergolide treatment. |
|
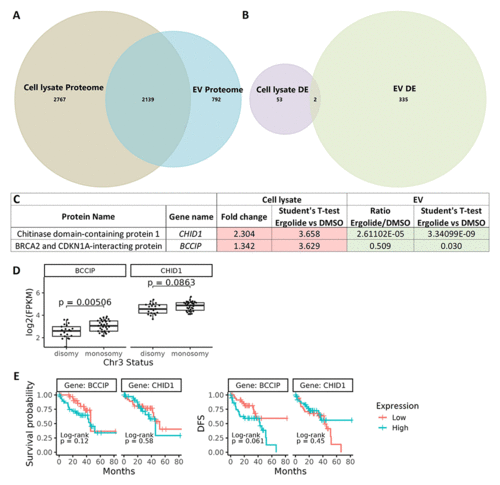
Comparison of cellular lysate and EV proteome. (A) Venn diagram depicting 2139 common proteins in the proteome of OMM2.5 cell lysate and OMM2.5 EV samples. (B) Among the differentially expressed proteins, two common proteins were identified in the OMM2.5 cell lysate and EV samples. (C) Table presenting CHID1 and BCCIP as significantly upregulated (red) in ergolide treated OMM2.5 cell lysate and downregulated (green) in EVs isolated from ergolide treated OMM2.5 cells. (D) Higher BCCIP (p = 0.005) expression levels noted in chromosome 3 (Chr 3) monosomy UM patients, with no difference in CHID1 expression level and Chr 3 status. (E) High or low expression levels of BCCIP and CHID1 did not correlate to overall survival or disease-free survival (DFS) in UM TCGA patient samples. |