- Title
-
Tissue-specific temperature dependence of RNA editing levels in zebrafish
- Authors
- Li, W., Bu, M., Hu, R., Jiang, S., Chen, L., Somero, G.N.
- Source
- Full text @ BMC Biol.
|
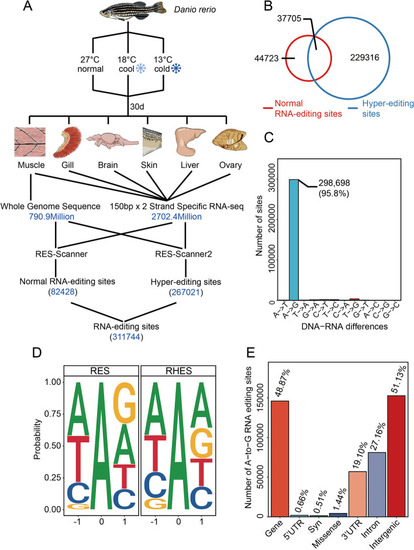
Experimental strategies and the RNA editome in zebrafish. |
|
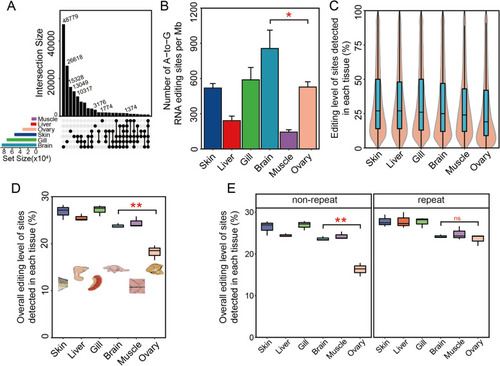
Characteristics of tissue-specific editomes in zebrafish. |
|
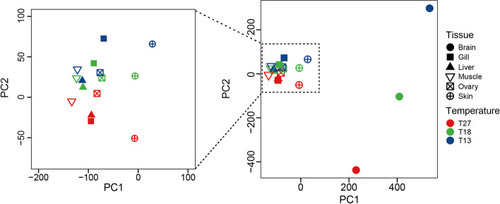
Principal component analysis of RNA editing across tissues from zebrafish acclimated to different temperatures |
|
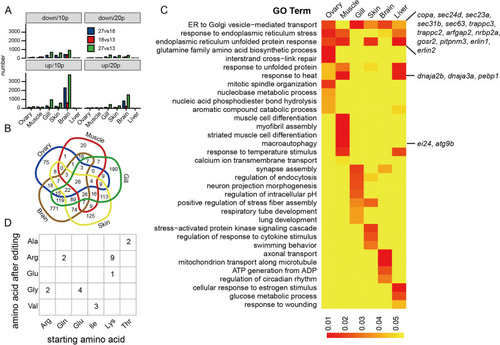
Differential A-to-I RNA editing sites and their related function. |
|
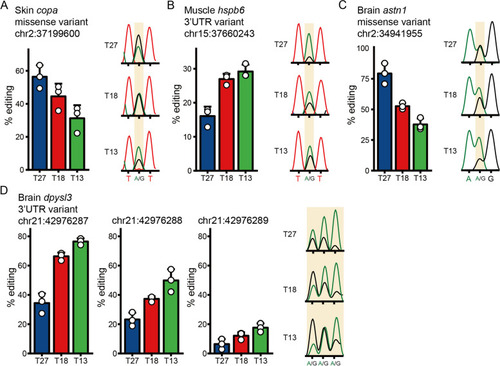
Validation of the dynamic change of RNA editing sites by Sanger sequencing. For each panel, the left bar graph indicates the editing level detected by the in silico method for the three temperatures (Data are shown as the mean ± SD, |
|
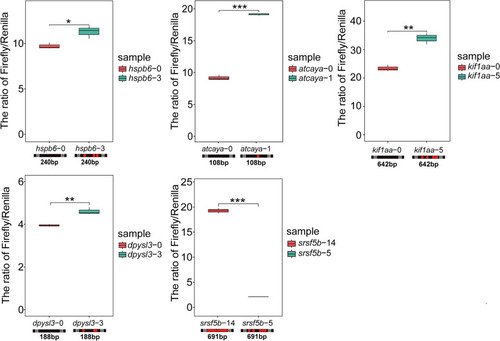
The relationship between the number of edited sites and efficiency of protein synthesis. The ratio of firefly (test) to |