- Title
-
Identification of ancestral gnathostome Gli3 enhancers with activity in mammals
- Authors
- Ali, S., Abrar, M., Hussain, I., Batool, F., Raza, R.Z., Khatoon, H., Zoia, M., Visel, A., Shubin, N.H., Osterwalder, M., Abbasi, A.A.
- Source
- Full text @ Dev. Growth Diff.
|
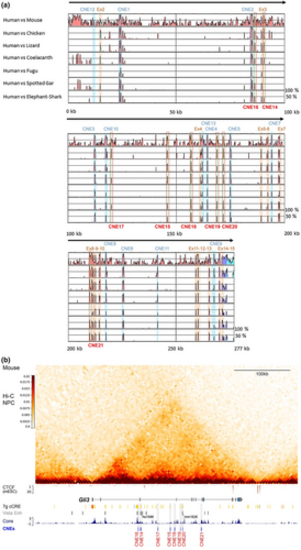
Comparative genomic analysis reveals deeply conserved GLI3 intronic CNEs with associated enhancer identity. (a) Multispecies sequence alignment of the genomic interval containing the human GLI3 |
|
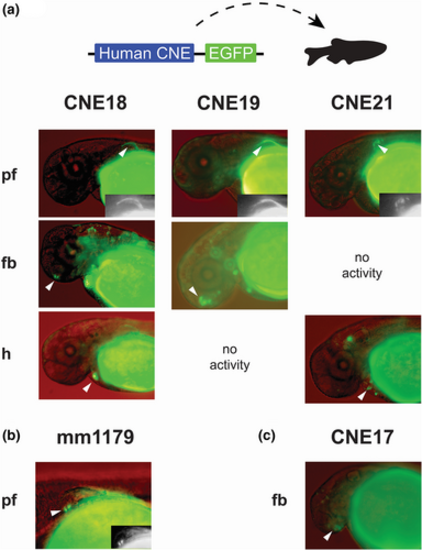
Gli3–CNEs conserved in slowly evolving fish genomes exhibit enhancer activity in Gli3-related tissues. (a) CNEs with highest sequence identity between human and elephant shark drive reproducible GFP transgenic reporter activity in multiple zebrafish embryonic tissues, including the pectoral fin (pf), forebrain (fb), and heart (h). (b) The mm1179 mouse embryonic Gli3 limb enhancer was validated in zebrafish for comparison and drives restricted activity in the pf. (c) CNE17 promotes transcriptional activity exclusively in the fb. Representative images (merged bright field and fluorescent signals) of live zebrafish transgenic embryos are shown and white arrowheads indicate reproducible GFP reporter activities (see also Figure S2 and Table S3). Inlets for elements active in the pf show GFP-only signals (in grayscale). Orientation of embryos is anterior to the left and dorsal to the top, with a lateral view. |
|
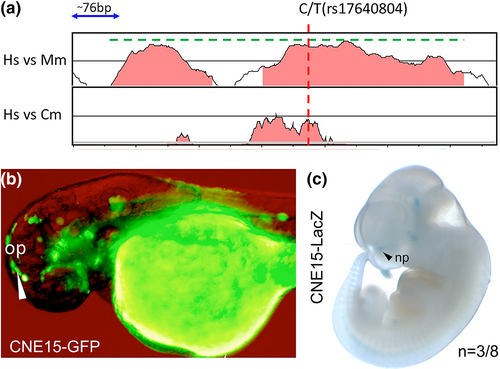
Implication of a GLI3–CNE in disease-related sequence variation. (a) VISTA plot showing CNE15 deep conservation down to elephant shark (cm; Callorhinchus milii) using human as a baseline. Human–mouse conserved sequence is shown underneath the dashed green line (566 bp) and is extended by ~50 bp in both directions to cover the human–mouse conserved peak included for transgenesis. (b) CNE15 enhancer induced transgenic GFP reporter expression in the olfactory placode of zebrafish embryos at 48 hpf (white arrowhead). (c) CNE15 drives LacZ reporter activity in the presumptive upper nasal process (np) at the boundary to the forebrain in mouse embryos at E11.5 (black arrowhead). “n” indicates the number of transgenic mouse embryos with LacZ activity in the np. op, olfactory placode; np, nasal process. |
|
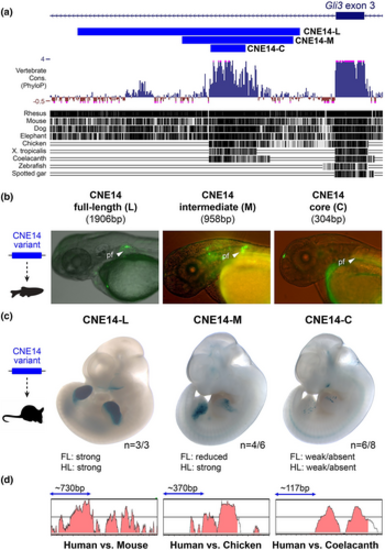
CNE extension correlates with evolutionary gain of enhancer function. (a) Genomic location and conservation of CNE14 variants based on UCSC PhyloP and MultiZ alignment tools. CNE14 human–mouse conserved region (CNE-L, 1906 bp), CNE14 intermediate human–chicken region (CNE-M, 958 bp), and the core conserved human–coelacanth sequence (CNE-C, 304 bp) are compared. (b) CNE14-L and CNE14-M induce GFP expression in the developing zebrafish fish pectoral fin. (c) Transgenic LacZ reporter activities of CNE14 versions in mouse embryos at E11.5. While CNE14-L induced strong LacZ expression in the anterior limb mesenchyme, activity driven by CNE14-M and CNE14-C was progressively reduced. “n” indicates the number of embryos with reproducible transgenic reporter activity versus the total number of transgenic embryos showing any LacZ signal. (d) Pairwise sequence comparison of CNE14 by Vista plots. Blue double arrow indicates scale. FL, forelimb; HL, hindlimb. |