- Title
-
Multivalent insulin receptor activation using insulin-DNA origami nanostructures
- Authors
- Spratt, J., Dias, J.M., Kolonelou, C., Kiriako, G., Engström, E., Petrova, E., Karampelias, C., Cervenka, I., Papanicolaou, N., Lentini, A., Reinius, B., Andersson, O., Ambrosetti, E., Ruas, J.L., Teixeira, A.I.
- Source
- Full text @ Nat Nanotechnol
|
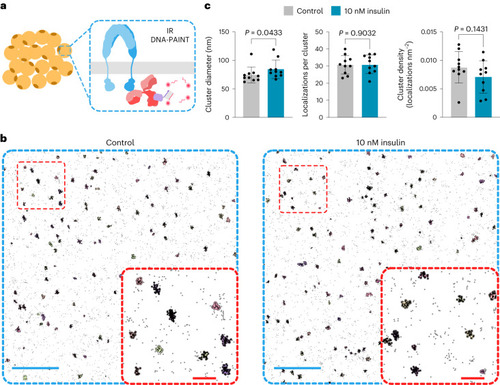
IRs are organized in nanoclusters at the cell membrane. |
|
Insulin NanoRods with programmable insulin configurations. |
|
Valency of insulin on NanoRods determines the residence time of IR binding. |
|
Valency and spacing of insulin on NanoRods determine IR pathway activation. |
|
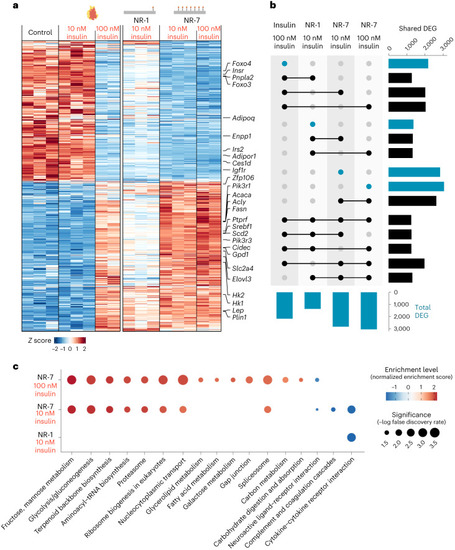
Valency of insulin on NanoRods modulates transcriptional responses. |
|
Valency of insulin on NanoRods determines their capacity to lower free glucose in β-cell-ablated zebrafish larvae. |