- Title
-
The prolyl isomerase Pin1 stabilizes NeuroD during differentiation of mechanoreceptors
- Authors
- Zhao, L., Fong, S.H., Yang, Q., Jiang, Y.J., Korzh, V., Liou, Y.C.
- Source
- Full text @ Front Cell Dev Biol
|
Sequence analysis of zebrafish Pin1. |
|
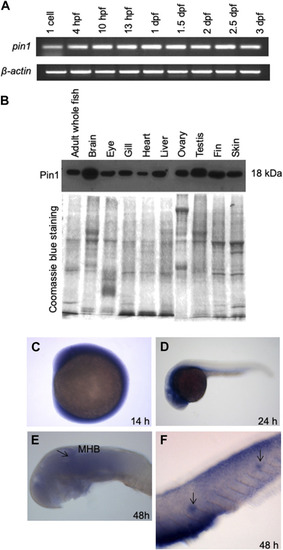
The spatial and temporal expression patterns of zebrafish Pin1. |
|
Zebrafish embryonic developmental delay in |
|
Specific PLL neuromasts hair cells defects in |
|
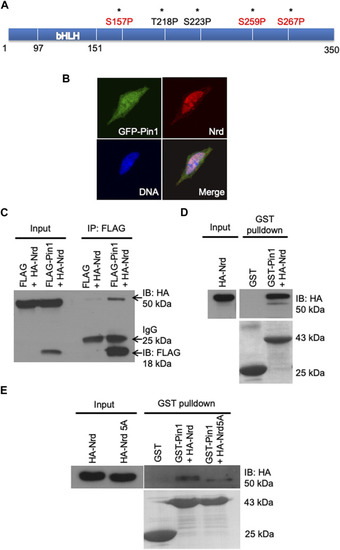
Zebrafish Pin1 interacts with Nrd via pSer/Thr-Pro motifs. |
|
Pin1 regulates Nrd stability. Protein stability assay of HA-Nrd in HEK 293T cells stably expressing control siRNA and Pin1 siRNA. |