- Title
-
Alternative Splicing and Nonsense-Mediated Decay of a Zebrafish GABA Receptor Subunit Transcript
- Authors
- Moisan, G.J., Kamath, N., Apgar, S., Schwehr, M., Vedmurthy, P., Conner, O., Hayes, K., Toro, C.P.
- Source
- Full text @ Zebrafish
|
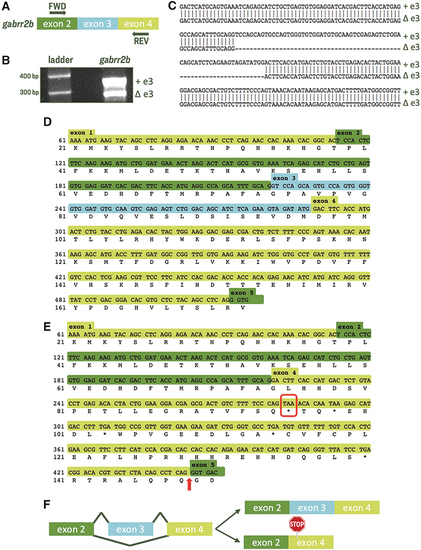
Exon 3 is alternatively spliced in zebrafish gabrr2b. (A) Schematic illustrating the location of FWD and REV PCR primers relative to GABAA receptor subunit ρ2b gene gabrr2b exon 3. (B) RT-PCR was performed on RNA extracted from ∼40 larvae from a single clutch of eggs. Agarose gel of PCR product shows two amplicons from gabrr2b amplification with primers flanking exon 3. (C) Sequence alignment of the two products gel-extracted from (B) reveal a novel isoform of ρ2b missing exon 3 (Δe3). (D, E) Translation of exons 1–5 of transcripts containing exon 3 (+e3; D), and missing exon 3 (Δe3; E). Comparison of the two amino acid sequences reveals a frameshift and PTC in exon 4 of Δe3-gabrr2b transcripts. Arrow highlights exon–exon junction 186 nt downstream of PTC. (F) Schematic representing AS outcomes. AS, alternative splicing; FWD, forward; PTC, premature termination codon; REV, reverse; RT-PCR, reverse transcription-polymerase chain reaction. |
|
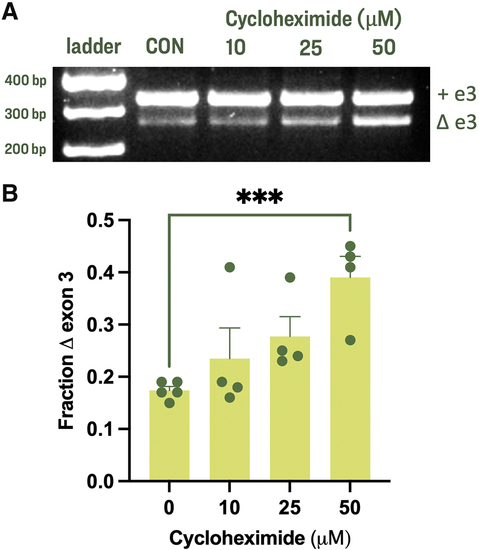
Inhibiting NMD results in a larger proportion of Δe3-gabrr2b mRNAs. (A) Representative gel from RT-PCR amplification around gabrr2b exon 3 from control larvae and those treated with 10, 25, or 50 μM of translation-inhibitor CHX for 24 h. (B) Average + SEM fraction of Δe3-gabrr2b transcripts as compared with +e3-gabrr2b transcripts as calculated from band intensities. n = 4–5 replicates per condition (shown as individual data points), with 40 larvae from a single clutch treated simultaneously before RT-PCR of each replicate. ***Control versus 50 μM CHX: p = 0.0006 (two-tailed unpaired t-test). CHX, cycloheximide; NMD, nonsense-mediated decay; SEM, standard error of the mean. |