- Title
-
Lipid droplets are a metabolic vulnerability in melanoma
- Authors
- Lumaquin-Yin, D., Montal, E., Johns, E., Baggiolini, A., Huang, T.H., Ma, Y., LaPlante, C., Suresh, S., Studer, L., White, R.M.
- Source
- Full text @ Nat. Commun.
|
Zebrafish melanoma displays distinct transcriptional states where melanocytic cell state upregulates oxidative metabolic pathways. |
|
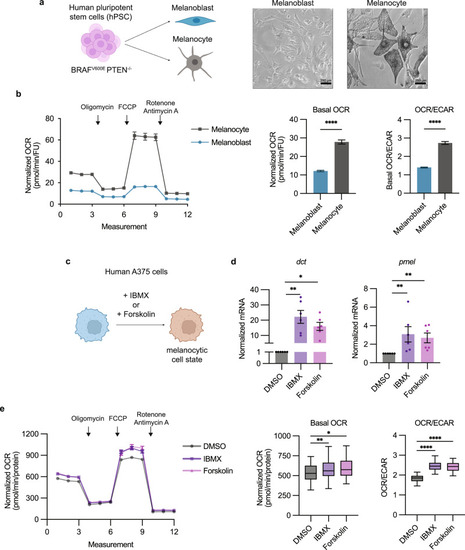
Melanocytic cell state leads to oxidative metabolic rewiring. |
|
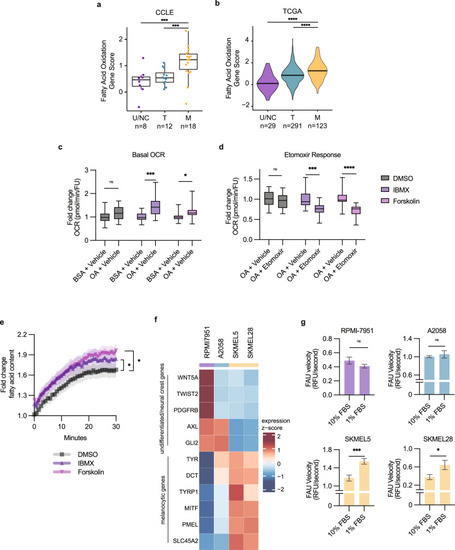
Fatty acids are utilized by melanocytic cells for oxidative metabolism. Fatty acid oxidation gene score for undifferentiated/neural crest (U/NC), transitory (T), and melanocytic (M) melanomas in the databases from: |
|
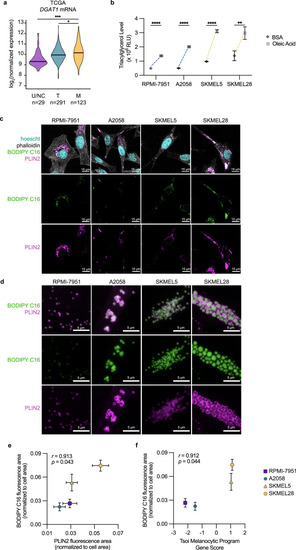
Melanocytic cells increase lipid droplet production. |
|
Knockout of DGAT1 suppresses lipid droplet formation and tumor progression. |
|
Inhibiting lipid droplet formation suppresses tumor growth and cell cycle progression. |