- Title
-
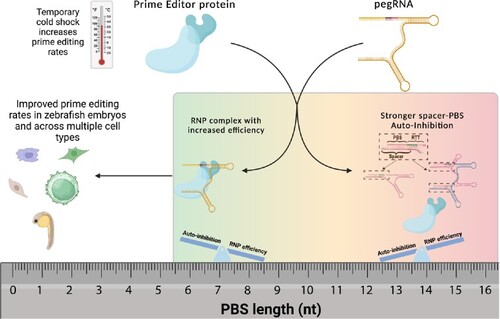
Reducing the inherent auto-inhibitory interaction within the pegRNA enhances prime editing efficiency
- Authors
- Ponnienselvan, K., Liu, P., Nyalile, T., Oikemus, S., Maitland, S.A., Lawson, N.D., Luban, J., Wolfe, S.A.
- Source
- Full text @ Nucleic Acids Res.
|
|
|
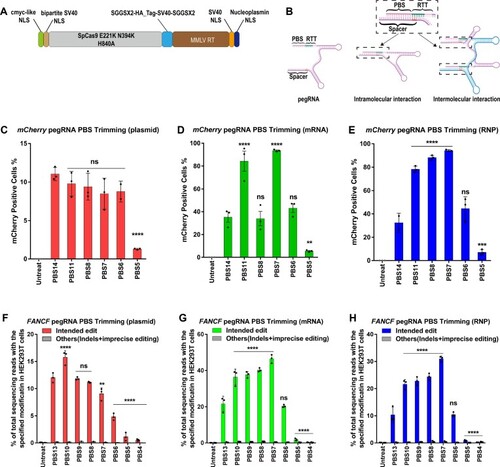
PE2 editing efficiency as a function of pegRNA PBS length at three genomic sites in HEK293T cells using three different delivery platforms. ( |
|
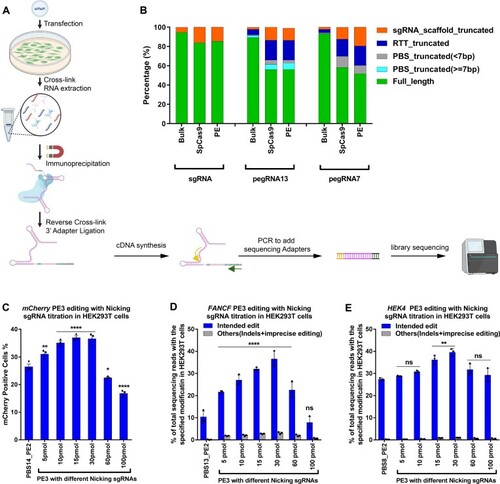
Small RNA-seq analysis of different pegRNA species bound to the Prime editor in HEK293T cells. ( |
|
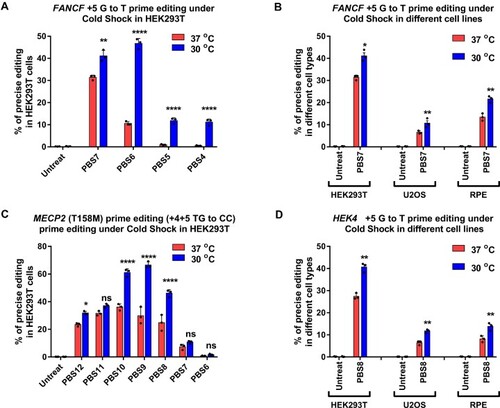
Cold shock enhances Prime editing rates in human cells. (A, C) RNP-mediated PE2 editing efficiency at the specified positions with varying pegRNA PBS length for ( |
|
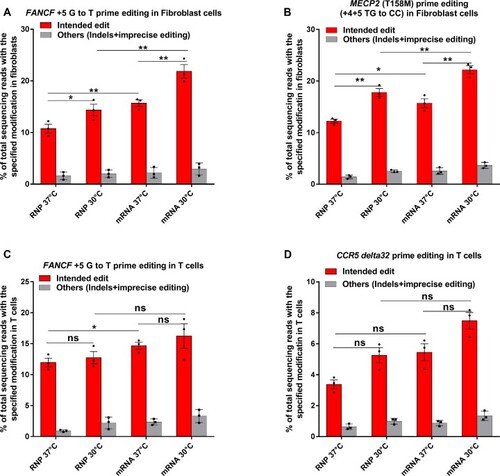
Prime editing efficiency in primary Fibroblasts and Primary T cells. (A, B) RNP and mRNA-mediated PE3 editing efficiencies at the specified positions for ( |