- Title
-
Molecular Physiological Evidence for the Role of Na+-Cl- Co-Transporter in Branchial Na+ Uptake in Freshwater Teleosts
- Authors
- Shih, S.W., Yan, J.J., Lu, S.W., Chuang, Y.T., Lin, H.W., Chou, M.Y., Hwang, P.P.
- Source
- Full text @ Int. J. Mol. Sci.
|
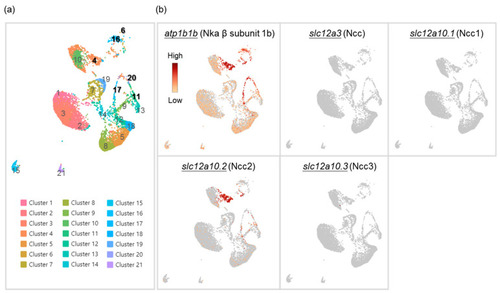
Cell clustering of gill cells by single-cell RNA sequencing (scRNA-Seq) analysis in adult zebrafish. scRNA-Seq shows 21 cell clusters in the gill on the UMAP (a). The distribution and expression of atp1b1b (Nka β subunit 1b), slc12a3 (Ncc), slc12a10.1 (Ncc1), slc12a10.2 (Ncc2), and slc12a10.3 (Ncc3) are shown on UMAP in the zebrafish gill cells (b). |
|
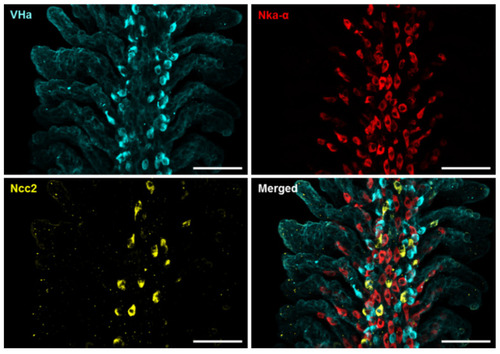
Localization of the three subtypes of ionocytes in the gills of adult zebrafish. Whole-mount immunofluorescence (IF) of the gills was used to reveal the expression patterns of VHa-rich (HR) ionocytes, Nka-rich (NaR) ionocytes, and NCC ionocytes with antibodies against vascular-type H+ ATPase (VHa), Nka α subunit (Nka-α), and Ncc2, respectively. A different angle of this image sample is shown in Figure S2. Scale bar, 50 μm. |
|
Effects of low-Na+ acclimation on Ncc2 expression in the gills of adult zebrafish. The mRNA expression levels of slc12a10.2 (Ncc2) in the gills were analyzed by quantitative real-time polymerase chain reaction (qRT-PCR) after acclimation to low-Na+ FW. qRT-PCR data were normalized to rpl13a (N = 7) (a). The protein expression of Ncc2 in the gills acclimated to low-Na+ FW was analyzed by Western blot. The blots show the bands with molecular weights corresponding to Ncc2 and GAPDH (approximately 112 kDa and 35 kDa, respectively). The protein expression of Ncc2 was quantified and normalized to GAPDH (N = 6) (b). Values are the mean ± SD. Student’s t-test, * p < 0.05. ns, not significant. |
|
Effects of low-Na+ acclimation on the number of NCC ionocytes and Ncc2 expression within individual NCC ionocytes in the gills of adult zebrafish. The cell number of NCC ionocytes and the fluorescence intensity of Ncc2 signals in the gill filaments were analyzed by whole-mount IF after acclimation to low-Na+ FW (a). NCC ionocyte number of filaments (N = 12) (b), mean fluorescence intensity of individual NCC ionocytes (N = 531–1017) (c), and the average mean fluorescence intensity of NCC ionocytes of filaments (N = 12) (d) are shown. Values are the mean ± SD or SEM. Student’s t-test or Mann–Whitney U test, *** p < 0.001, **** p < 0.0001. Scale bar, 50 μm. |
|
Effects of low-Na+ acclimation on branchial Na+ uptake capacity of adult zebrafish. The Na+ gradient at the gills of adult fish was measured using the scanning ion-selective electrode technique (SIET) after 7 d of acclimation to low-Na+ FW. Values are the mean ± SD (N = 8). Student’s t-test, **** p < 0.0001. |
|
Acute effects of Cl−-free FW on the branchial Na+ uptake capacity of adult zebrafish. Na+ gradients at low-Na+-acclimated gills were analyzed before and after the acute incubation of Cl−-free (a) or normal Cl− (b) FW media using the SIET. The dashed line represents the same fish. Values are the mean ± SD (N = 5–7). Student’s t-test, **** p < 0.0001. ns, not significant. |
|
Effects of metolazone on the branchial Na+ uptake capacity of adult zebrafish. Na+ gradients at low-Na+-acclimated gills were analyzed before and after the treatments of metolazone (a) or DMSO (b) using the SIET. The dashed line represents the same fish. Values are the mean ± SD (N = 5–6). Student’s t-test, *** p < 0.001. ns, not significant. WT, wild-type. |
|
Effects of EIPA on the branchial Na+ uptake capacity of adult zebrafish. Na+ gradients at low-Na+-acclimated gills were analyzed before and after the treatments of EIPA (a) or DMSO (b) using the SIET. The dashed line represents the same fish. Values are the mean ± SD (N = 5–7). Student’s t-test, **** p < 0.0001. ns, not significant. EIPA, 5-(N-ethyl-N-isopropyl)-amiloride. WT, wild-type. |