- Title
-
Novel Loss-of-Function Variant in HNF1a Induces β-Cell Dysfunction through Endoplasmic Reticulum Stress
- Authors
- Chen, Y., Jia, J., Zhao, Q., Zhang, Y., Huang, B., Wang, L., Tian, J., Huang, C., Li, M., Li, X.
- Source
- Full text @ Int. J. Mol. Sci.
|
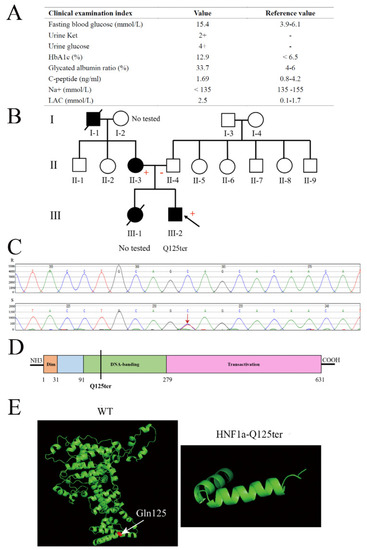
Clinical and functional characteristics of the HNF1a-Q125ter variant. ( |
|
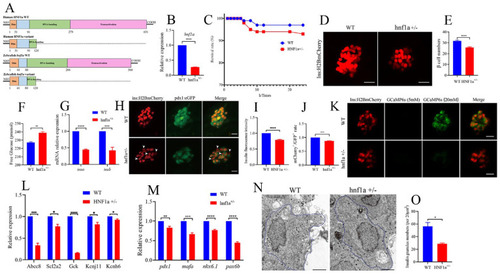
Similar variant impaired β-cell function in zebrafish. ( |
|
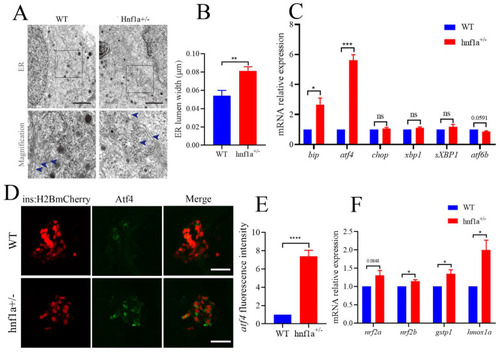
The β cells of |
|
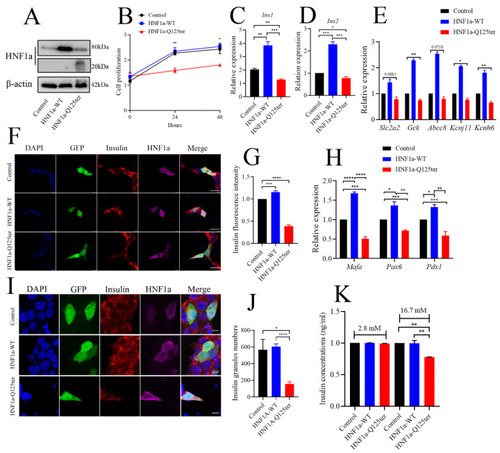
HNF1a-Q125ter impaired β-cell function in vitro. ( |
|
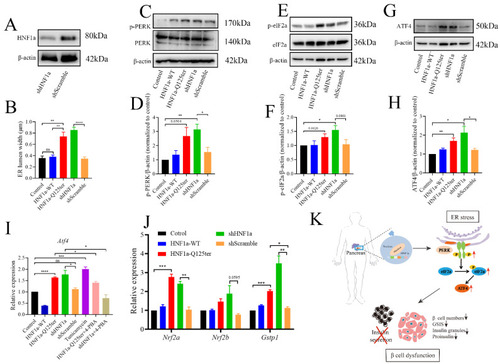
HNF1a-Q125ter induced ER stress through PERK/eIF2a/ATF4 signaling pathway. ( |