- Title
-
Investigation of the causal etiology in a patient with T-B+NK+ immunodeficiency
- Authors
- Sertori, R., Lin, J.X., Martinez, E., Rana, S., Sharo, A., Kazemian, M., Sunderam, U., Andrake, M., Shinton, S., Truong, B., Dunbrack, R.M., Liu, C., Srinivasan, R., Brenner, S.E., Seroogy, C.M., Puck, J.M., Leonard, W.J., Wiest, D.L.
- Source
- Full text @ Front Immunol
|
Identification of the |
|
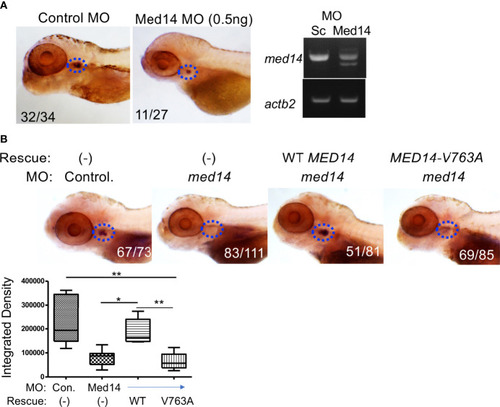
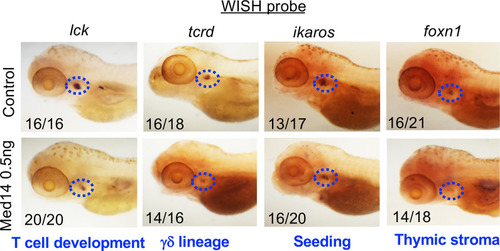
Role of Med14 in Zebrafish T cell Development. |
|
Role of MED14 in Development of Thymic Subpopulations in Zebrafish. The effect of |
|
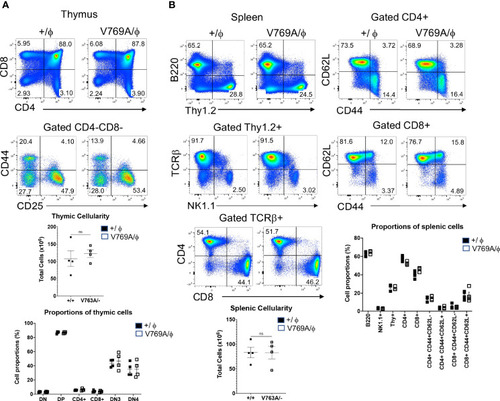
Phenotypic analysis of lymphoid development in |
|
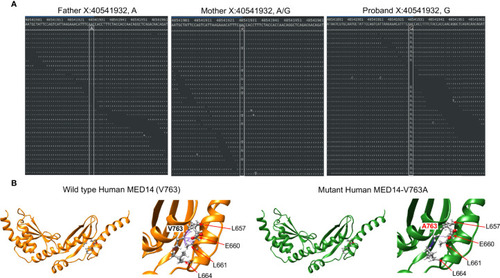
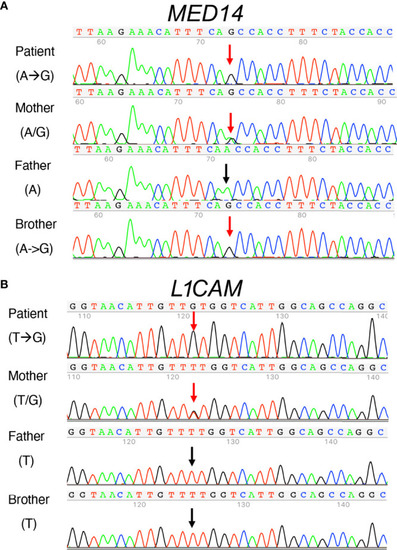
Sanger sequence analysis of MED14 V763A and L1CAM Q498H variants in the patient, his parents, and his healthy brother. |
|
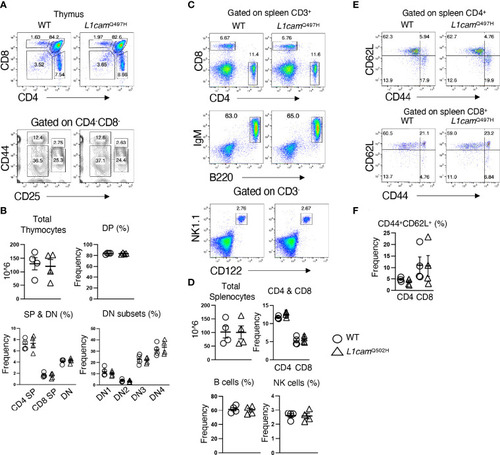
Phenotypic analysis of the |

ZFIN is incorporating published figure images and captions as part of an ongoing project. Figures from some publications have not yet been curated, or are not available for display because of copyright restrictions. |